Abstract
Understanding the relationship between gene diversity and function for important environmental processes are major ecological research goals. We applied gene-targeted-metagenomics and pyrosequencing to aromatic dioxygenase genes to obtain greater sequence depth than possible by other methods. A PCR primer set designed to target a 524 bp region that confers substrate specificity of biphenyl dioxygenases yielded 2000 and 604 sequences from 5′ and 3′ ends of the PCR products, respectively, that passed our validity criteria. Sequence alignment showed three known conserved residues as well as another seven conserved residues not previously reported. Ninety-five and 41% of the valid sequences were assigned to 22 and 3 novel clusters in that they did not include any previously reported sequences at 0.6 distance by Complete Linkage Clustering for the sequenced regions. The greater diversity revealed by this gene-targeted approach provides deeper insights into genes potentially important in environmental processes to better understand their ecology, functional differences and evolutionary origins. We also provide criteria for primer design for this approach as well as guidance for data processing of diverse functional genes since gene databases for most genes of environmental relevance are limited.
Keywords: Biphenyl, Dioxygenase, Gene diversity, Metagenomics, Pyrosequencing
Introduction
Metagenomics circumvents the problem of unculturability and has the potential to understand microbial communities at their aggregate level, transcending the individual organism to focus on the genes in a community (National Research Council, 2007). Because of the extensive genetic diversity of most microbial communities, it is currently impossible to obtain enough sequence depth to sample any gene with sufficient coverage to draw meaningful conclusions about its diversity or population characteristics. This is particularly important where functional screens are problematic due to expression requirements such as for protein complexes. To overcome this limitation, approaches are needed to target sequencing capacity to genes of particular interest. One approach is to use polymerase chain reaction (PCR)-based targeting together with pyrosequencing technology, similar to how this is currently done for 16S rRNA gene sequencing (Sogin et al., 2006; Huber et al., 2007). This approach, which we term gene-targeted-metagenomics (GT-metagenomics), should then provide more extensive insight into the diversity that nature has produced as well as provide sequence information for probes to use in recovering the entire gene of clades of interest. Since many studies have shown that the mutation of a few amino acids can critically affect the structure of individual enzymes, changing their substrate utilization and degradation activities (e.g. Parales et al., 2000; Suenaga et al., 2002; Bagneris et al., 2005; Vardar and Wood, 2005), understanding gene diversity in the nature can reveal functional, ecological and evolutionary patterns for key genes.
The GT-metagenomics approach requires that the targeted gene has sufficiently conserved regions of appropriate distance for emulsion PCR, which is required for pyrosequencing, so that primers or sets of primers will sufficiently cover a gene family. This approach is likely to be most useful for genes directly responsible for important ecosystem functions or ecological processes such as biogeochemical cycles, biodegradation, pathogenesis, antibiotic resistances, and cell signaling. We have tested this approach on a set of dioxygenase genes important in the carbon cycle for turnover of more recalcitrant organic carbon as well as for pollutant degradation. We also discuss requirements for primer design for GT-metagenomics and the subsequent data analysis. Our results show a much greater diversity of genes potentially important in nature’s carbon cycle that previously realized.
Materials and methods
Sample soil and DNA extraction
Polychlorinated biphenyl (PCB) contaminated soil (15 mg/kg) was collected from the root zone of an Austrian pine (Pinus nigra) tree at the grounds of a paint production plant in the Czech Republic that was previously shown to have significantly higher numbers of PCB degraders (Leigh et al., 2006). The DNA was extracted as described previously (Leigh et al., 2007) and stored at −20 °C until use.
Primer design
Gibson and Parales (2000) classified Rieske non-heme iron dioxygenase genes into four families; toluene/biphenyl, naphthalene, benzoate and phthalate. The Functional Gene Pipeline/Repository (FGPR) (http://fungene.cme.msu.edu/) provided the database of the sets of genes that belong to those families based on monthly Hidden Markov Model (HMM) (Durbin et al., 1998) searches of the DDBJ/EMBL/GenBank database. Nucleotide and protein sequences of the toluene/biphenyl family dioxygenase genes were retrieved from FGPR bph v3.1 database (July 2007) with a cutoff score >900 and size >400bp. The 40 retrieved protein sequences with 35 different nucleotide sequences were aligned using ClustalW (Thompson et al., 1994). A PCR primer set was designed from the conserved regions of these sequences: BPHD-f3, 5′-AACTGGAARTTYGCIGCVGA-3′; BPHD-r1, 5′-ACCCAGTTYTCICCRTCGTC-3′. The specificity of the primer set was tested by comparison with the DDBJ/EMBL/GenBank database.
Pyrosequencing
PCR primers with sequencing adapter A (5′-GCCTCCCTCGCGCCATCAG-3′) or B (5′-GCCTTGCCAGCCCGCTCAG-3′) at the 5′ end of BPHD-f3 or BPHD-r1 were synthesized and purified by dual HPLC (Integrated DNA Technologies, Coralville, IA). The PCR mixture was prepared in a total volume of 20 μl, containing 1 X FastStart High Fidelity Reaction Buffer (Roche Diagnostics, Basel, Switzerland), 1.25 μM of each primer, 150 ng μl−1 of bovine serum albumin (New England BioLabs), 0.2 mM of dNTPs, 0.5 μl (2.5 U) of FastStart High Fidelity PCR System Enzyme Blend (Roche Diagnostics) and 4 ng of template DNA. The PCR condition was optimized using the genomic DNA of Burkholderia xenovorans LB400 which carries one of the target dioxygenase genes. Amplifications were performed as follows: 3 min at 95 °C followed by 30 cycles of 45 sec at 95 °C, 45 sec at 60 °C and 40 sec at 72 °C then final extension for 4 min at 72 °C. PCR products with primer pairs A-BPHD-f3 and B-BPHD-r1 or B-BPHD-f3 and A-BPHD-r1 were sequenced from 5′ and 3′ ends, respectively. Triplicate PCR products with predicted 542 bp fragments were purified with QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany) then with QIAquick PCR Purification Kit (QIAGEN). DNA concentration was determined using NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Purified PCR products were mixed together with other bar-coded samples and subjected to pyrosequencing using Genome Sequencer FLX System (454 Life Sciences, Branford, CT).
Determination of valid sequences
The sequences were first trimmed of the primer region and low quality sequences were removed as follows. We plotted the number of sequences against the first position of an ambiguous base call or stop codon. We determined the length for analysis to be at the position where the plotted curve dropped sharply (175 bp for BPHD-f3 sequences and 200 bp for BPHD-r1 sequences). To remove possible frameshift errors, each sequence was used as a BLASTX query against a BLAST database of the translated library using a 0.001 E-value cutoff to determine the top 10 closest matches. Those reads with the greatest number of hits that include any out-of-frame segments were successively discarded (as both query and subject), until all out-of-frame results had been removed. Only the sequences that passed this filter were used for further data analysis and are termed as obtained sequences.
Conservation analysis
Four hundred and sixty-seven reference sequences, which have both the Rieske family domain (Pfam PF00355) and the Ring_hydroxyl_A family domain (Pfam PF00848), were retrieved from the Pfam protein family database (Finn et al., 2008). Deduced amino acid sequences from our obtained sequences were aligned using MUSCLE (Edgar, 2004) together with the corresponding region of the reference protein sequences. Gap-treated Shannon entropy (H′) (Zhang et al., 2007) at each alignment position was calculated as:
where fi,a is the relative frequency of amino acid a at the alignment position i. fi,gap represents the number of gaps at the alignment position i divided by the number of alignment sequences.
Distance calculation and clustering
Dissimilarity matrices were calculated from the individual pairwise alignments of amino acid sequences using MUSCLE with the default setting. Each value in the matrix is the fraction of the number of positions with dissimilar amino acids out of the total number of alignment positions compared excluding the end gaps. Dissimilar amino acid pairs were those for which negative probability values were assigned in BLOSUM62. These matrices were fed to DOTUR (Schloss and Handelsman, 2005) for Complete Linkage Clustering.
Nucleotide sequence accession numbers
The nucleotide sequences described in this study have been submitted to European Read Archive under the accession number of ERA000082.
Results
Primer design
The BPHD-f3 and BPHD-r1 primer set, corresponding to 219N – 225E and 387D – 393V of bphA1 from B. xenovorans LB400, respectively, was designed to amplify 524 bp PCR products. It includes a portion of the C-terminal domain of the large oxygenase subunit that is highly diverse and has many residues responsible for congener selectivity of PCBs (Vézina et al., 2008), including four regions identified by Mondello et al. (1997). With this primer set, theoretically 31 genes out of 49 genes from bph v4.9 (FGPR, January, 2009) with a cutoff score >900 and size >400bp can be amplified and no non-dioxygenase genes can be amplified using the BLAST search against the DDBJ/EMBL/GenBank database. When we tried to amplify the remaining 18 genes by increasing degeneracy of the primer set, several weak non-target bands were observed on agarose gels which might produce non-target sequences by pyrosequencing (see also discussion). These results indicated high specificity and sufficient coverage by the designed primer set. The predicted 524 bp PCR products were successfully amplified from genomic DNA of B. xenovorans LB400 and Rhodococcus sp. RHA1 which both carry target dioxygenase genes and not from genomic DNA of Sphingomonas wittichii RW1 which carries dioxin dioxygenase, that was not used for primer design.
Pyrosequencing statistics
Since the average sequence length produced by the Genome Sequencer FLX System is around 200–250 bp (Rothberg and Leamon, 2008), we sequenced the same pool of PCR products from both 5′ (BPHD-f3 sequences) and 3′ ends (BPHD-r1 sequences) (Table 1). Although the concentration of the sequenced PCR products for BPHD-f3 and BPHD-r1 were almost the same, the number of obtained sequences differed almost three fold, possibly because of the different efficiency of emulsion PCR caused by the different primer sequences attached to the adapter. Primer-trimmed sequence length varied from 28 to 293 bp. Out of over 3000 sequences, we determined 2632 sequences (79.3%) as obtained sequences for further analysis based on our position of error and frameshift analysis. The rarefaction curves at 0, 0.1, 0.2, 0.3, 0.4, 0.5 and 0.6 clustering distances are shown in Supplementary Figure S1.
Table 1.
Pyrosequencing information.
| Sequencing primer |
|||
|---|---|---|---|
| BPHD-f3 | BPHD-r1 | Sum | |
| Number of raw sequences | 2486 | 835 | 3321 |
| Total raw sequence length | 556 744 bp | 198 152 bp | 754 896 bp |
| Average raw sequence length | 224 bp | 237 bp | |
| Obtained sequencea length | 175 bp (58 aa) | 200 bp (66 aa) | |
| Number of obtained sequences | 2024 | 608 | 2632 |
| Number of valid sequencesb | 2000 | 604 | |
| Unique nucleotide sequences in valid sequences | 743 | 339 | |
Sequences that passed position of error and frameshift analysis
The number of sequences which have 230D, 233H and 239H for BPHD-f3 alignment and 344P for BPHD-r1 alignment. (The number correspond to the position of bphA1 from B. xenovorans LB400.
Conserved regions
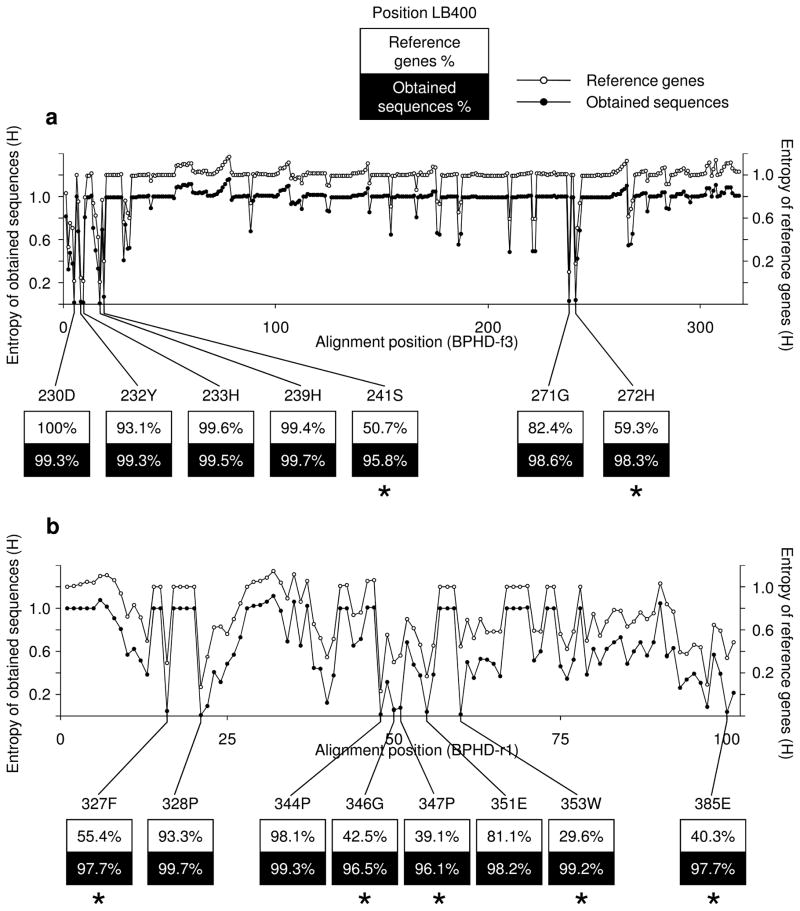
We examined the conservation of residues among obtained sequences and reference sequences by calculating the Shannon entropy, H′, at each alignment position. Overall patterns of conservation indicated by negative peaks of H′, are similar between obtained sequences and reference sequences (Figure 1). Seven and eight highlighted residues in BPHD-f3 and BPHD-r1 alignments, respectively, were identical among more than 95% of either obtained sequences or reference sequences (Figure 1). 230D, 232Y, 233H, 239H, 271G, 328P, 344P, and 351E which were conserved among more than 80% of reference sequences were also highly conserved among more than 98% of the obtained sequences. On the contrary, 241S and 272H in BPHD-f3 alignment and 327F, 346G, 347P, 353W and 385E in BPHD-r1 alignment were highly conserved among obtained sequences, but they were poorly conserved among reference sequences (29.6–59.3% of the reference sequences). Based on the crystal structure study, 241S, 353W and 385E are located on alpha-helix 8, beta-strand 19, and alpha-helix 14, respectively. None of the seven residues except 241S is a part of the active site, interface residues, or substrate binding pocket (Furusawa et al., 2004).
Figure 1.
Shannon entropy (H′) at each alignment position and conserved residues among obtained sequences and/or reference sequences for (a) BPHD-f3 sequences and (b) BPHD-r1 sequences. Open circles (○) indicate entropy of reference sequences and filled circles (●) indicate entropy of obtained sequences. The corresponding position numbers and the residues of bphA1 from B. xenovorans LB400 are indicated. The ratio of residues conserved in either set with >95% are shown. The residues highly conserved only among obtained sequences are indicated with [*].
We validated our obtained sequences using the conserved residues. The highly conserved 230D, 233H and 239H (Figure 1a) are located in fragments forming the substrate-binding pocket (Furusawa et al., 2004). 233H and 239H have been reported as residues to coordinate the mononuclear iron of Rieske non-heme iron dioxygenase and 230D plays a role of a bridge connecting the two iron sites, 123H and 233H (Furusawa et al., 2004; Dong et al., 2005). There are 24 sequences (1.2% of the obtained sequences of BPHD-f3) that did not have one or more of those three residues, indicating they were possible non-functional dioxygenase genes. Seventeen of them showed similarity with E-values of less than 10−4 to putative dioxygenase genes from environmental samples by BLASTx. The rest of the sequences either showed similarity to non-dioxygenase genes or less similarity (E-value of larger than 10−4) to any of the genes in the database. Since those sequences were missing functionally important residues, they could be caused by non-specific PCR amplification or evolutionary degeneracies in nature. For the BPHD-r1 side, since we could not find any common structural information of the protein for this region among Rieske non-heme iron dioxygenase genes, we used highly conserved 344P (Figure 1b) for validation of the sequences. Four sequences (0.7% of the obtained sequences of BPHD-r1) did not have this conserved residue. All of them showed similarity with E-values of less than 10−4 to putative dioxygenase genes from environmental samples by BLASTx. We excluded those 24 and 4 sequences that did not have the highly conserved and functionally important residues from further clustering and distribution analysis. Of the remaining 2604 sequences, which we termed as valid, 2075 sequences (80%) have highest BLASTx hits to dioxygenase genes (either cultured or environmental) with E-values of <10−4. The remaining 529 sequences (521 sequences from BPHD-f3 and 8 sequences from BPHD-r1) have no hits with E-values of <10−4 to any of the sequences in the database. The fact that those sequences have the highly conserved residues suggests they are novel dioxygenase genes.
Clustering and distribution of the obtained sequences
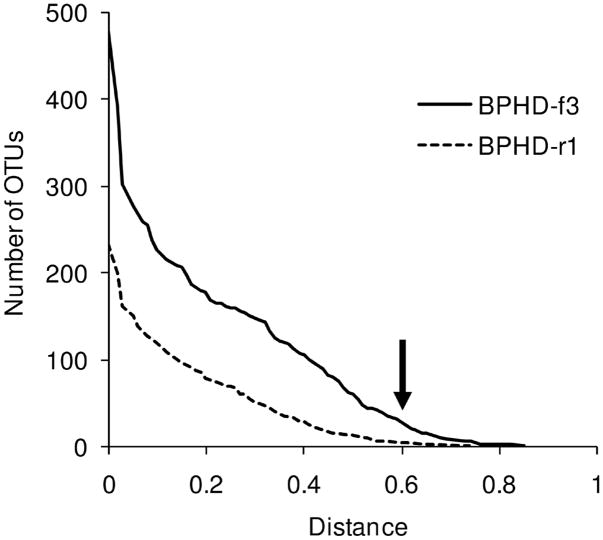
Those 2604 valid sequences with conserved positions were clustered together with reference sequences by Complete Linkage Clustering based on amino acid sequences. The numbers of operational taxonomical units (OTUs) of valid sequences at each distance level are shown in Figure 2. Four-hundred seventy-nine and 234 unique sequences were obtained from BPHD-f3 and BPHD-r1, respectively (Figure 2).
Figure 2.
Clustering of valid sequences at different distance levels by Complete Linkage Clustering based on amino acid sequences. The numbers of OTUs of valid sequences (solid line, BPHD-f3 sequences; dashed line, BPHD-r1 sequences) at each distance are shown. The arrow indicates the distance level used for the distribution analysis.
Since pyrosequencing produces a large dataset, it is necessary to cluster the sequences to reduce the number for analysis. We selected 0.6 distance as a cutoff (Figure 2) to obtain 56 and 20 clusters for BPHD-f3 and BPHD-r1, respectively, numbers manageable for analysis and interpretation. Figure 3 shows the distance to the closest reference sequence(s) for each sequence in each cluster and the number of sequences in that cluster. The clusters are (i) newly obtained clusters composed of only our valid sequences (closed symbols in Figure 3) and previously known clusters composed of (ii) both our valid sequences and reference sequences (open symbols in Figure 3), and (iii) only reference sequences. The numbers of clusters in each category are (i), (ii), (iii): 22, 11, 23 and 3, 4, 13 for BPHD-f3 and BPHD-r1, respectively. Fifty-nine percent of the sequences from BPHD-f3 are in the largest four clusters which were type (i). The rest of the sequences are distributed into small clusters which were composed of less than 6% (120 sequences) of the total valid sequences. The largest type (ii) cluster from BPHD-f3 is composed of 35 sequences, (1.8%). The cluster includes toluene/biphenyl dioxygenase genes such as bphA1 from B. xenovorans LB400 (AAB63425) and bphAa from Rhodococcus sp. RHA1 (ABG99107) is the second largest type (ii) cluster with 18 sequences (0.9%). Three sequences from BPHD-f3 had a similarity value of 1 to the reference sequences. For BPHD-r1 sequences, 85% of the valid sequences were in the largest two clusters and the rest were in small clusters which have less than 8% (48 sequences) of the total valid sequences. The largest cluster has 288 sequences (48%) and was type (ii). The second largest cluster is type (i) and has 223 sequences (37%). None of the sequences from BPHD-r1 had a similarity value of 1 to the reference sequences. The farthest distance to the reference sequences was 0.59 and most sequences in novel clusters have distances between 0.45–0.6 to reference sequences (Figure 3). Cluster distribution and frequency of each unique sequence for the clusters with more than 200 sequences are shown in Supplementary Figure S2. The accession numbers of reference sequences in each cluster are shown in Supplementary Table S1.
Figure 3.
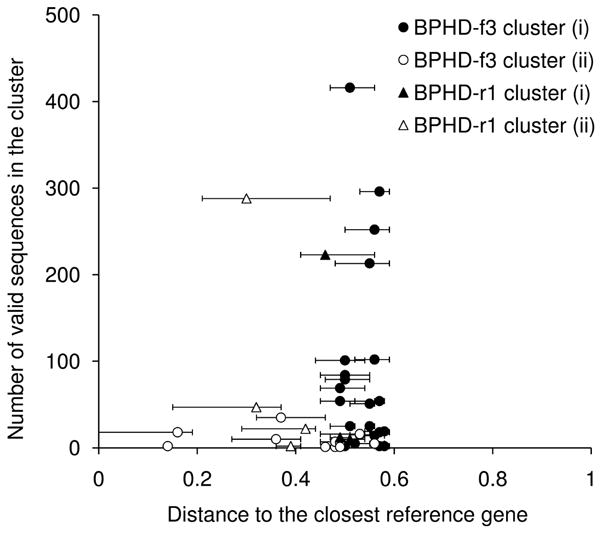
Pairwise distance to the closest reference sequence(s) for each valid sequence and the number of sequences in each cluster. Symbols are the median of the distances in the cluster. Error bar indicates the range of the distance.
Discussion
The particular region amplified, degree of coverage of the target gene family and the specificity of primer sets are critical for designing PCR primer sets to assess gene populations in a community. For pyrosequencing, the length of the PCR product must not be too long or it will reduce emulsion PCR efficiency (Margulies et al., 2005). We selected the particular 524 bp region because it provided suitably conserved primer sites, was of suitable length and covered a region known to be functionally important (confers substrate specificity). Our primer set had 48- and 16-fold degeneracy for BPHD-f3 and BPHD-r1, respectively, to obtain a good coverage of the target genes. Since increasing degeneracy caused less specificity of the primer set, the degeneracy used here was optimal for maximizing the coverage without sacrificing specificity. After we removed low quality sequences (likely due to sequencing error and/or non-dioxygenase sequences) and those without conserved positions, 80% and 72% of the raw sequences remained as valid sequences for BPHD-f3 and BPHD-r1, respectively. These high ratios indicate enough primer specificity for further analysis. As a result, 95% and 41% of the valid sequences were assigned to novel (type (i)) clusters. Although we designed the primers using only toluene/biphenyl dioxygenase, it is interesting that the deeper sequencing allowed us to obtain a much broader range of apparent dioxygenase genes. For example, clusters F24 and R5 contain all well-known toluene/biphenyl dioxygenase genes and clusters F35 and R7 contain all well-known naphthalene dioxygenase genes (Figure S2 and Table S1). This provides a perspective on the size of the clusters with and without reference sequences and hence on the diversity of the novel dioxygenase genes.
The detection of novel conserved residues, such as the seven found in this study, illustrates one of the useful outputs from the GT-metagenomics approach, in that it better reveals what nature has selected that can not be seen from either pure culture or shotgun metagenomics approaches. Based on previous reports, six out of the seven residues have not been studied for their structural or functional importance for Rieske non-heme iron dioxygenase genes such as biphenyl dioxygenase (Furusawa et al., 2004), cumene dioxygenase (Dong et al., 2005), or naphthalene dioxygenase (Karlsson et al., 2003). However, this high conservation ratio suggests that they play some structural or functional role and hence provide new targets for site-directed mutagenesis studies. Moreover, those novel conserved regions allow us to design new primer sets to explore further diversity of the genes.
The larger challenge in GT-metagenomics is the data analysis. We tested several approaches. First, we selected a representative sequence from each 0.6 distance cluster based on amino acid sequences and tried to build a phylogenetic tree. However, those sequences were too diverse to produce a reliable tree (the bootstrap values were very low). Then, we tried a model approach to show the distribution of the pyrosequenced sequences against known genes. The model approach is used to search and create databases based on the relatedness to a certain group of functional genes such as by FGPR. We built two protein models based on a set of known dioxygenase genes in the toluene/biphenyl family and a set of all well-known Rieske non-heme iron dioxygenase genes, which includes toluene/biphenyl, naphthalene, benzoate and phthalate families. The valid sequences were searched against those models using HMM and scores of each sequence to the model were calculated. Only 68% of the valid sequences had scores to either of the models for BPHD-f3 sequences. In contrast, 100% of the valid BPHD-r1 sequences had scores to the either of the models. This might be because the model was based on known genes and could not give similarity scores to the completely novel genes. The advantage of this method is that we can compare the sequences based on the score to the model regardless of the region and length. When there is enough reference gene information, e.g. by adding new clusters obtained in this and other studies, to provide a more stable model, the model approach will be a useful method to map and compare sequences obtained from different regions of the gene.
The most abundant clusters in BPHD-r1 sequences were the novel (type (i)) cluster with 48% of the total valid sequences (Figure 3). However, there is no such large type (i) cluster in BPHD-f3. In the same manner, we found differences between BPHD-f3 and BPHD-r1 clustering in terms of proportion of the clusters in each type. This suggests further complexity of environmental dioxygenases composed of different combinations of those clusters in one gene. Advances in sequencing technology that provide greater read length, such as the recently introduced GS FLX Titanium Series, which can extend read length to 400–500 bp (http://www.454.com/) will be a great help with GT-metagenomics approaches.
Since this GT-metagenomics approach is based on PCR products, it like all PCR-based approaches should be assumed to have primer bias and cannot be comprehensive or reflect actual quantification. It can however reveal much more sequence information about the genes it does recover and hence more insight into what nature has produced. Functional genes often have less conserved regions and the current sequenced length is less than desired. Thus using several different primer sets, which can be designed from new conserved regions, and combining information from the model approach should prove for better understanding the biology behind gene diversity. This information should also provide the probes or other tools that can aid the recovery of full-length gene sequences from the environment for functional studies. Also, testing the expression and activity of those genes toward aromatic hydrocarbons will be important to suggest their function in nature.
Supplementary Material
Acknowledgments
We thank Chike V. Anadumaka for technical assistance, and acknowledge the important work by Mary Beth Leigh, Martina Mackova, Tomas Macek, and Ondrej Uhlik, which established this rhizosphere site as important for aromatic biodegradation studies. This study was supported by NIEHS grant under the Superfund Basic Research Program 5P42 ES004911–18/19.
Footnotes
Supplementary information is available at The ISME Journal’s website.
References
- Bagnéris C, Cammack R, Mason JR. Subtle difference between benzene and toluene dioxygenases of Pseudomonas putida. Appl Environ Microbiol. 2005;71:1570–80. doi: 10.1128/AEM.71.3.1570-1580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XS, Fushinobu S, Fukuda E, Terada T, Nakamura S, Shimizu K, et al. Crystal structure of the terminal oxygenase component of cumene dioxygenase from Pseudomonas fluorescens IP01. J Bacteriol. 2005;187:2483–2490. doi: 10.1128/JB.187.7.2483-2490.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R, Eddy S, Krogh A, Mitchison G. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press; 1998. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Nagarajan V, Tanokura M, Masai E, Fukuda M, Senda T. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J Mol Biol. 2004;342:1041–1052. doi: 10.1016/j.jmb.2004.07.062. [DOI] [PubMed] [Google Scholar]
- Gibson DT, Parales RE. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr Opin Biotechnol. 2000;11:236–243. doi: 10.1016/s0958-1669(00)00090-2. [DOI] [PubMed] [Google Scholar]
- Huber JA, Mark Welch D, Morrison HG, Huse SM, Neal PR, Butterfield DA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Karlsson A, Parales JV, Parales RE, Gibson DT, Eklund H, Ramaswamy S. Crystal structure of naphthalene dioxygenase: Side-on binding of dioxygen to iron. Science. 2003;299:1039–1042. doi: 10.1126/science.1078020. [DOI] [PubMed] [Google Scholar]
- Leigh MB, Pellizari VH, Uhlik O, Sutka R, Rodrigues J, Ostrom NE, et al. Biphenyl-utilizing bacteria and their functional genes in a pine root zone contaminated with polychlorinated biphenyls (PCBs) ISME J. 2007;1:134–148. doi: 10.1038/ismej.2007.26. [DOI] [PubMed] [Google Scholar]
- Leigh MB, Prouzová P, Macková M, Macek T, Nagle DP, Fletcher JS. Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl Environ Microbiol. 2006;72:2331–2342. doi: 10.1128/AEM.72.4.2331-2342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondello FJ, Turcich MP, Lobos JH, Erickson BD. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl Environ Microbiol. 1997;63:3096–3103. doi: 10.1128/aem.63.8.3096-3103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. The New Science of Metagenomics. The National Academies Press; Washington DC: 2007. Why Metagenomics? pp. 12–32. [Google Scholar]
- Parales RE, Lee K, Resnick SM, Jiang H, Lessner DJ, Gibson DT. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J Bacteriol. 2000;182:1641–1649. doi: 10.1128/jb.182.6.1641-1649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg JM, Leamon JH. The development and impact of 454 sequencing. Nat Biotechnol. 2008;26:1117–1124. doi: 10.1038/nbt1485. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga H, Watanabe T, Sato M, Ngadiman, Furukawa K. Alteration of regiospecificity in biphenyl dioxygenase by active-site engineering. J Bacteriol. 2002;184:3682–3688. doi: 10.1128/JB.184.13.3682-3688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardar G, Wood TK. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for enhanced chlorinated ethene degradation and o-xylene oxidation. Appl Microbiol Biotechnol. 2005;68:510–517. doi: 10.1007/s00253-005-1923-4. [DOI] [PubMed] [Google Scholar]
- Vézina J, Barriault D, Sylvestre M. Diversity of the C-terminal portion of the biphenyl dioxygenase large subunit. J Mol Microbiol Biotechnol. 2008;15:139–151. doi: 10.1159/000121326. [DOI] [PubMed] [Google Scholar]
- Zhang SW, Zhang YL, Pan Q, Cheng YM, Chou KC. Estimating residue evolutionary conservation by introducing von Neumann entropy and a novel gap-treating approach. Amino Acids. 2007;35:495–501. doi: 10.1007/s00726-007-0586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.