Abstract
The fusion oncoproteins PAX3-FOXO1 [t(2;13)(q35;q14)] and PAX7-FOXO1 [t(1;13)(p36;q14)] typify alveolar rhabdomyosarcoma (ARMS); however, 20-30% of cases lack these specific translocations. In this study, cytogenetic and/or molecular characterization to include FISH, RT-PCR and sequencing analyses of five rhabdomyosarcomas [four ARMS and one embryonal rhabdomyosarcoma (ERMS)] with novel, recurrent t(2;2)(p23;q35) or t(2;8)(q35;q13) revealed that these non-canonical translocations fuse PAX3 to NCOA1 or NCOA2 respectively. The PAX3-NCOA1 and PAX3-NCOA2 transcripts encode chimeric proteins composed of the paired-box and homeodomain DNA-binding domains of PAX3, and the CID domain, the Q-rich region and the AD2 domain of NCOA1 or NCOA2. To investigate the biological function of these recurrent variant translocations, the coding regions of PAX3-NCOA1 and PAX3-NCOA2 cDNA constructs were introduced into expression vectors with tetracycline-regulated expression. Both fusion proteins showed transforming activity in the soft agar assay. Deletion of the AD2 portion of the PAX3-NCOA fusion proteins reduced the transforming activity of each chimeric protein. Similarly, but with greater impact, CID domain deletion fully abrogated the transforming activity of the chimeric protein. These studies: (1) expand our knowledge of PAX3 variant translocations in RMS with identification of a novel PAX3-NCOA2 fusion; (2) show that both PAX3-NCOA1 and PAX3-NCOA2 represent recurrent RMS rearrangements; (3) confirm the transforming activity of both translocation events and demonstrate the essentiality of intact AD2 and CID domains for optimal transforming activity; and, (5) provide alternative approaches (FISH and RT-PCR) for detecting PAX-NCOA fusions in nondividing cells of RMS. The latter could potentially be utilized as aids in diagnostically challenging cases.
INTRODUCTION
Rhabdomyosarcomas are the largest subset of soft tissue sarcomas in infants and children and are highly heterogeneous, clinically aggressive tumors that show varying degrees of skeletal muscle differentiation (Raney et al., 2001). The two main histological patterns recognized include embryonal rhabdomyosarcoma (ERMS) and alveolar rhabdomyosarcoma (ARMS). Morphologic evaluation alone is often insufficient to make the distinction between ARMS and ERMS as some ARMS lack the alveolar architecture (“solid variant”) and ERMS can be densely cellular and poorly differentiated (Parham et al., 2006). This distinction is clinically critical however, in assigning patients appropriately to high-risk therapeutic regimens. Thus, a valuable diagnostic adjunct in ARMS is the identification of translocations t(2;13)(q35;q14) and t(1;13)(p36;q14), and the associated PAX3-FOXO1 and PAX7-FOXO1 fusion transcripts, respectively. Recognition of these specific translocations is also prognostically important as PAX3-FOXO1 positive ARMSs are significantly more aggressive than PAX7-FOXO1 ARMSs (Sorensen et al., 2002).
The vast majority of ARMSs exhibit PAX-FOXO1 fusion transcripts that encode for proteins composed of a PAX paired domain and homeodomain DNA-binding element (NH2-terminus) and a FOXO1 transactivation domain (COOH-terminus) (Galili et al., 1993; Davis et al., 1994). The PAX-FOXO1 chimeric products are highly expressed in the tumor cells, exclusively localized in the nucleus, and are more potent transcriptional activators than wild type PAX3, PAX7 or FOXO1. Approximately 30% of ARMSs fail to exhibit either PAX3-FOXO1 or PAX7-FOXO1 fusions by routine RT-PCR and have been termed “fusion transcript negative” (Barr et al., 2002). Rare, isolated cases have shown alternate translocation associated fusion transcripts (PAX3-AFX and PAX3-NCOA1) (Barr et al., 2002; Wachtel et al., 2004). In the current study, a novel genetic subgroup of rhabdomyosarcoma with recurrent chromosomal translocations fusing PAX3 to members of the nuclear receptor coactivator family of genes to include NCOA1 [t(2;2)(p23;q35)] or NCOA2 [t(2;8)(q35;q13)] in lieu of a forkhead domain family member was identified and characterized.
MATERIALS AND METHODS
Tumor Samples
Five rhabdomyosarcoma specimens that were determined to be negative for a rearrangement of the FOXO1 locus by FISH or a PAX3- or PAX7-FOXO1 fusion transcript by RT-PCR were included in the study. The clinicohistopathologic features of the patients and corresponding tumors are listed in Table 1. The histopathological diagnoses were established in accordance with the International Classification of Rhabdomyosarcoma criteria (Qualman et al., 1998). Histologic and immunohistochemical features typical of ARMS and ERMS were identified in the respective cases (Fig. 1). No distinctive clinical phenotype was recognized that might distinguish these PAX-NCOA fusion positive cases from other alveolar or embryonal rhabdomyosarcomas. The karyotype of one patient has been reported previously (case 5) (Meloni-Ehrig et al., 2009).
Table 1.
Clinicopathologic, Cytogenetic, and Fusion Gene Data
| Case | Agea/ Sex |
Locationb (size in cm) |
Histopathologic Diagnosisc |
Karyotyped | Fusion Genee |
|---|---|---|---|---|---|
| 1 | 22/M | Base of skull (2.0 × 1.3 × 1.1) |
ARMS | NP | PAX3-NCOA1 |
| 2 | 5/M | Gluteus maximus N/A |
ARMS | 46,XY[15] | PAX3-NCOA1 |
| 3 | 18/F | Perineum N/A |
ARMS | NP | PAX3-NCOA1 |
| 4 | 2/M | Testis (5.0 × 2.5 × 2.3) |
ARMS | 66-90,XY,add(X)(p11)×2, t(2;8)(q35;q12)×2[cp12]/46,XY[3] |
PAX3-NCOA2 |
| 5 | 2wk/ M |
Perineum (3.7 × 4.1 × 3.2) |
ERMS | 46,XY,t(2;8)(q35;q13)[6]/46,XY[14] f | PAX3-NCOA2 |
Age is in years unless otherwise stated
All cases were primary lesions
ARMS, alveolar rhabdomyosarcoma; ERMS, embryonal rhabdomyosarcoma
N/A, not available; NP, not performed
Fusion gene status for each case as confirmed by FISH and RT-PCR with subsequent sequencing
This karyotype has been reported previously (Meloni-Ehrig et al., 2009)
Figure 1.
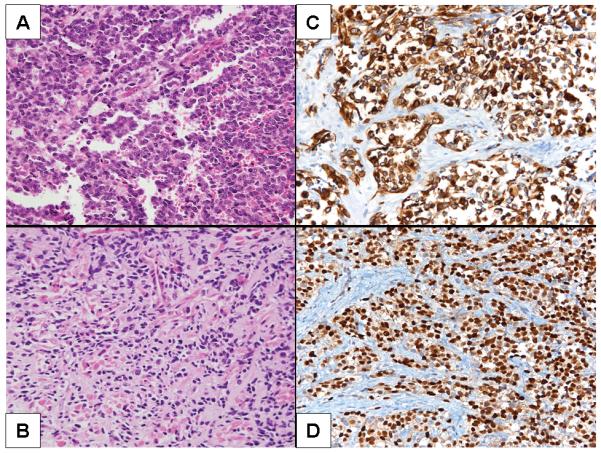
A. Alveolar rhabdomyosarcoma, Case 1. Note the loss of cellular cohesion. B. Scattered rhabdomyoblasts with eosinophilic cytoplasm in Case 5, embryonal rhabdomyosarcoma. C and D. Case 4, desmin and myogenin immunoreactivity, respectively.
Cytogenetic Analysis
Cytogenetic analysis was performed on sterile representative samples of cases 2, 4, and 5 using standard culture and harvesting procedures (Bridge et al., 2000). Metaphase cells were banded with Giemsa trypsin, and the karyotypes were described according to the International System for Human Cytogenetic Nomenclature 2009 (ISCN, 2009).
Probe Design and Development
Bacterial artificial chromosome (BAC) and yeast artificial chromosome (YAC) clones for the PAX3, NCOA1, and NCOA2 gene regions were identified utilizing the NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/mapview), the Ensembl Genome Browser (http://www.ensembl.org), the University of California Santa Cruz (UCSC) Human Genome Browser Gateway (http://genome.ucsc.edu/cgi-bin/hgGateway), and the Whitehead Institute/MIT Center Genome Browser (http://www-genome.wi.mit.edu/cgi-bin/contig/yac_info). The PAX3 probe sets utilized have been previously described (Nishio et al., 2006). Combinations of probe sets were custom-designed to flank and span NCOA1 and NCOA2 (Table 2).
Table 2.
Optimized NCOA1 and NCOA2 FISH Probe Sets
| Probe set | Clonea | Location | Label |
|---|---|---|---|
| RP11-715M22 | BAC | Proximal to NCOA1 locus | Spectrum green |
| RP11-24J3 | BAC | Distal to NCOA1 locus | Spectrum orange |
| RP11-1109B14/RP11-803B18 | BAC cocktail | Spans NCOA1 locus | Spectrum green |
| CTD-2606A20/RP11-68N15 | BAC cocktail | Proximal to NCOA2 locus | Spectrum green |
| RP11-680D10/RP11-133E24 | BAC cocktail | Distal to NCOA2 locus | Spectrum orange |
| RP11-479K21/RP11-183D21 | BAC cocktail | Spans NCOA2 locus | Spectrum green |
BAC, bacterial artificial chromosome
Fluorescence In Situ Hybridization
Two-color FISH studies were performed on cytologic touch preparations (n = 2), in situ metaphase cell preparations (n = 1), and formalin-fixed, paraffin-embedded tissue sections (n = 3). Probes were directly labeled by nick translation with either Spectrum Green or Spectrum Orange-dUTP utilizing a modification of the manufacturer's protocol (Abbott Molecular Inc., Des Plaines, IL, USA) (Nishio et al., 2006; Kapels et al., 2007).
Prior to hybridization, the touch preparations and in situ slides were pretreated as previously described (Nishio et al., 2006). Following pretreatment, the cells and probes were codenatured at 75°C for 1-6 min and incubated at 37°C overnight using the HYBrite™ denaturation/hybridization system (Nishio et al., 2006; Kapels et al., 2007). Post-hybridization washing was performed in 0.4x SSC/0.3% NP-40 at 72°C for 2 min, followed by 2x SSC/0.1% NP-40 at room temperature for 1 min. The slides were air-dried in the dark and counterstained with 10μl of 4′,6-diamidino-2-phenylindole (DAPI II; Abbott Molecular). Hybridization signals were assessed in 200 interphase nuclei with strong and well-delineated signals by two different individuals. An interphase cell specimen was interpreted as abnormal if spanning probe signals for the NCOA1 or NCOA2 genes fused with the spanning probe signal for the PAX3 gene, or if a split of flanking probe signals (defined as more than a signal size apart) was detected in more than 10% of the cells evaluated (more than two standard deviations above the average false-positive rate). Negative controls included normal peripheral blood lymphocytes and cytologic touch preparations of pathologically unremarkable skeletal muscle. Images were acquired by use of the CytoVision Image Analysis System (Applied Imaging, Santa Clara, CA, USA).
Identification of PAX3-NCOA1 and PAX3-NCOA2 FusionTranscripts
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA). The 3′-RACE (3′-rapid amplification of cDNA ends) was performed using the SMART-RACE cDNA amplification kit and protocol (Clontech, Palo Alto, CA). Briefly, first-strand cDNA was reverse transcribed from 0.5 μg total RNA using Superscript II and the 3′-RACE cDNA synthesis primer (3′-CDS) from the kit. An aliquot of the first-strand cDNA was then amplified using a PAX3 gene-specific forward primer (PAX3-32, Table 3) and a universal primer mix (SMART-RACE kit). Polymerase chain reaction (PCR) conditions were as recommended by the manufacturer. A nested PCR reaction using the nested universal primer (SMART-RACE kit) as the reverse primer and PAX3-34 (internal to PAX3-32, Table 3) as the forward primer was performed according to the manufacturer's instructions. Second-round PCR products were electrophoresed, purified, and subcloned into E. coli OneShot® competent cells (Invitrogen, Carlsbad, CA). Plasmid DNAs were purified, analyzed for the presence of insert by restriction enzyme digestion and sequenced.
Table 3.
Oligonucleotides Used in RACE and RT-PCR Studies
| Gene | Sequence | Position of the Primer1 |
|---|---|---|
| PAX3-32 | GCTTTCAACCTCTCTTCCCG | exon 6; 853-874 n |
| PAX3-34 | CCAGCCCACATCTATTCCACAAG | exon 7; 936-958 n |
| NCOA1-33 | TAATACATCTAATCCACCCCCCTG | exons 13-14; 2883-2906 n |
| NCOA1-35 | GCCTTCTCATCATTTCTCCCTTCTACTG | exon 13; 2764-2792 n |
| NCOA1-40; RACE | CCATCCTGTCTGTCTCACATAAGCC | exon 13; 2743-2767 n |
| PAX3-41 | TGGAGCGTGCTTTTGAGAGAAC | exon 5; 701-722 n |
| PAX3-43 | TCCTTCCAACCCAGACAGCAG | exon 7; 1032-1052 n |
| PAX3-45; RACE | CTGCCTCCCCAGCACCAG | exon 7; 1062-1079 n |
| PAX3-47; RACE | GCTTTCAACCATCTCATTCCCG | exon 6; 864-874 n |
| PAX3-42; RACE | ATGGCAGTGGGAGGGAACC | exon 7; 1244-1263 n |
| NCOA2-44 | CGGTGCCCATTCTCCAGATG | exon 13; 3011-3030 n |
| NCOA2-46; RACE | GCAGTGCTGTTTTCTGGGCTC | exon 12; 2732-2752 n |
| NCOA2-48 | TCTCACAGCCGAACTCTGCG | exon 13; 3029-3048 n |
RT-PCR for PAX3-NCOA1 and PAX3-NCOA2
First-strand cDNA was reverse transcribed from 0.5μg of total RNA with the Advantage RT-PCR System with random or oligo(dT) primers (Clontech). For the detection of a PAX3-NCOA1 fusion transcript, RT-PCR was conducted with a forward primer (PAX3-32, Table 3) specific for exon 6 of PAX3 and reverse primers (NCOA1-33 and NCOA1-35, Table 3) specific for different exons of NCOA1. Similarly, RT-PCR was used to detect PAX3-NCOA2 fusion transcripts, with PAX3 specific forward primers (PAX3-41, -45, Table 3) and NCOA2 specific reverse primers (NCOA2-44; -48, Table 3) the amplification products were cloned in pCR4/TOPO (Invitrogen), sequenced, and analyzed using MacVector Version 10 (MacVector Inc. Cary, NC).
Construction of Full Length PAX3-NCOA1 and PAX3-NCOA2 cDNA
Full length PAX3-NCOA1 and PAX3-NCOA2 cDNAs were constructed using cloned cDNA fragments obtained from the RT-PCR and RACE assays and inserted in pCR4-TOPO. The full length cDNA of PAX3-NCOA1 and PAX3-NCOA2 was assembled by PCR-based gene synthesis using a 2-step assembly/amplification protocol (Carr et al., 2004) with the exception that the assembly PCR was run for 45 cycles. The primary structure of the constructs was confirmed by nucleotide sequence analysis using pCR4 and gene specific sequencing primers.
Construction of Tetracycline Inducible, PAX3-NCOA1 or PAX3-NCOA2 Mammalian Expression Vectors
The coding region of PAX3-NCOA1 and PAX3-NCOA2 fusion constructs was PCR amplified using pCR4-TOPO cloned PAX3-NCOA1 and PAX3-NCOA2 with the Expand High Fidelity PCR System (Roche Diagnostics, Indianapolis, IN) in the presence of gene specific primers (Table 3). The amplified coding regions were introduced into the tetracycline inducible mammalian expression vector pcDNA4/TO, after sequencing two clones were selected; pcDNA4-PAX33-NCOA1.23 and pcDNA4-PAX3-NCOA2.1.
To establish the tetracycline-regulated system, NIH3T3 cells were first stably transfected using Effectene Transfection Reagent (Qiagen) with pcDNA6/TR (Invitrogen, Carlsbad, CA), which encodes the Tet repressor (TeR) under the control of human CMV promoter. Subsequently, clones were selected and maintained in tetracycline-free medium (Clontech) containing 5 μg/mL Blasticidin (Invitrogen). The inducibility of each clone was tested by transient transfection with pcDNA4/TO/LacZ and stained for ß-gal (Invitrogen). The clone (NIH 3T3-Tet-19.1) demonstrating the highest LacZ induction was selected for generating a stable cell line with tetracycline regulated PAX3-NCOA1 or PAX3-NCOA2 expression.
Construction of PAX3-NCOA1 or PAX3-NCOA2 Deletion Mutants
PAX3-NCOA1 or PAX3-NCOA2 deletion mutants in pcDNA4/TO were created using two consecutive steps to delete either AD1 or AD2 regions of NCOA1 and NCOA2 utilizing the QuickChange kit (Stratagene, La Jolla,CA) with primers listed in Table 4.
Table 4.
Oligonucleotides Used in the Construction of PAX3-NCOA1 and PAX3-NCOA2 Deletion Mutants
| Gene | Primer | Sequence |
|---|---|---|
|
PAX3- NCOA1 type 1 |
AD1-PN1.1-357u | GCTATAAATCAGAGTAAATCAGCTCTTGAACAGCTGGTATCC′ |
| AD1-PN1.1-357l | GGATACCAGCTGTTCAAGAGCTGATTTACTCTGATTTATAGC | |
| AD1-PN1.1-417u | GCTATAAATCAGAGTAAATCGGTGGATTAGATGTATTATCAGAG | |
| AD1-PN1.1-417l | CTCTGATAATACATCTAATCCACCGATTTACTCTGATTTATAGC | |
| AD2-PN1.1-697-727u | GTTCCCCAAGGTGAGGCCAACGCCTCCGGGTATCAGACATGA | |
| AD2-PN1.1-697-727l | TCATGTCTGATACCCGGAGGCGTTGGCCTCACCTTGGGGAAC | |
|
PAX3- NCOA1 type 2 |
AD1-PN1.2-495u | GAAGACCAGTGTATTAGCGAGAAGGCTTCTTCTTGA |
| AD1-PN1.2-495l | TCAAGAAGAAGCCTTCTCGCTAATACACTGGTCTTC | |
| AD1-PN1.2-549u | GAAGACCAGTGTATTAGCGGTGGATTAGATGTATTAATCAG | |
| AD1-PN1.2-549l | CTGATTAATACATCTAATCCACCGCTAATACACTGGTCTTC | |
| AD2-PN1.2-835-865u | GGTGAGGCCAACTTTGCTCCATCTCTAGACATGAAGGCCTGGCAGCAA | |
| AD2-PN1.2-835-865l | TTGCTGCCAGGCCTTCATGTCTAGAGATGGAGCAAAGTTGGCCTCACC | |
|
PAX3- NCOA2 |
AD1-PN2-649u | GCCAGCCAAAACAGGCAGCCAGACCAGGTGTATCTGGCCTTGCGG |
| AD1-PN2-649l | CCGCAAGGCCAGATACACCTGGTCTGGCTGCCTGTTTTGGCTGGC | |
| AD1-PN2-739u | GCCAGCCAAAACAGGCAGCCAGACCAGGTAGCTATTCGCCCATGCAAGATCCA | |
| AD1-PN2-739l | TGGATCTTGCATGGGCGAATAGCTACCTGGTCTGGCTGCCTGTTTTGGCTGGC | |
| AD2- PN2-898-93u | CAGCAGTTTCCATTTCCTCCAAACTACCCAACAGTCTCAGGCCAACCCAGCC | |
| AD2- PN2-898-931 | GGCTGGGTTGGCCTGAGACTGTTGGGTAGTTTGGAGGAAATGGAAACTGCTG |
u= upper strand primer; l=lower strand primer
Sequence Analysis
Plasmid clones were sequenced using ABI PRISM® BigDye™ Terminator Cycle Sequencing Kit version 2 and ABI PRISM® 3730 DNA Analyzer, a capillary electrophoresis system (ABI). Sequence analysis was performed using the MacVector with Assembler Version 10.0.2 (MacVector, Inc. Cary, NC) sequence analysis program.
Tetracycline Regulated Expression
The expression constructs pcDNA4-PAX33-NCOA1.23 and pcDNA4-PAX3-NCOA2.1 were transfected into NIH3T3-Tet-19.1 and selected on 500-μg/ml zeocin for 7 - 10 days. Tetracycline-free medium containing 5 μg/mL Blasticidin (Invitrogen) and 400 μg/mL Zeocin (Invitrogen) was used for maintaining the transfected cells. Induction of pcDNA4/-PAX3/NCOA1 and pcDNA4/TO-PAX3/NCOA2 and deletion mutants was accomplished by adding 1 μg/ml tetracycline to media and tested by reverse transcription-PCR (RT-PCR) and Western blot.
Growth of Cells in Semi-solid Media
Transformation of NIH3T3 cells by PAX3, PAX3-FOXO1, and their variants was examined using a colony formation in soft agar culture assay. Following selection, 4000 cells were embedded in soft agar as described by Lugo and Witte (1989) and were seeded in six well plates in culture medium (DMEM/F12) 14-21 days with fresh medium supplementation every 3 days before colonies were counted and photographed. Colonies larger than 100 mm were counted for each plate. Experiments were repeated three times with duplicate plates for each cell line.
Western Analyses and Immunoblotting
To verify the anticipated expression of the various constructs, the constructs were transfected into NIH 3T319.1 cells and expression of the cloned genes was induced by tetracycline (1 μg/ml). For detecting in vitro expression of PAX3-NCOA1 and PAX3-NCOA2 and their derivatives, confluent cells were rinsed with cold phosphate-buffered saline twice and lysed in the presence M-PER (Pierce, Thermo Fisher Scientific Inc. Rockford, IL). Whole cell lysates were fractionated on precasted 4-15% SDS-polyacrylamide gels (BioRad) and analyzed by Western blot. The expression of PAX3-NCOA1 and PAX3–NCOA2 or their derivatives was detected by the primary PAX3 antibody (N19, sc-7749, Santa Cruz Biotechnology, Inc. Santa Cruz CA). The secondary anti-rabbit IgG antibody was used at a dilution of 1:3000. To confirm that equal amounts of protein extract were loaded, the level of β-actin levels in the cell lysate were assessed using anti-β-actin (sc-81178) with secondary antibodies anti-goat IgG (sc-2378) and anti-mouse IgG (sc-2005), (Santa Cruz Biotechnology, Inc). Immune complexes were detected by chemiluminescence using SuperSignal reagents (Thermo Scientific, Rockford, IL).
RESULTS
Cytogenetic Analysis
Cytogenetic analysis conducted on cases 2, 4, and 5 revealed the presence of a balanced 2;8 translocation involving 2q35 and a near identical 8q breakpoint (q12-13) in cases 4 and 5 (Fig. 2). An abnormal clone was not identified in case 2.
Figure 2.
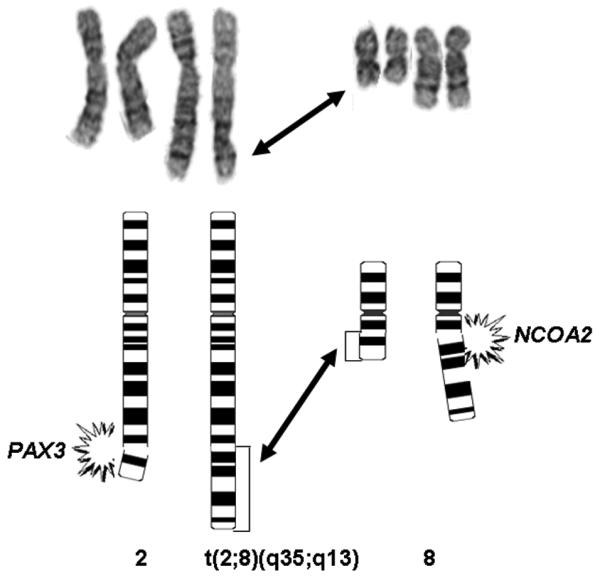
Partial karyotype and schematic illustrating the t(2;8)(q35;q12) identified in Case 4.
Fluorescence In Situ Hybridization
All cases exhibited a rearrangement of the PAX3 gene locus using the custom-designed breakpoint flanking probe set. Subsequent analysis with the custom-designed NCOA1 and NCOA2 breakpoint flanking and spanning probe sets confirmed the presence of a rearrangement of NCOA1 and fusion with PAX3 (PAX3-NCOA1) in cases 1, 2 and 3, and a rearrangement of NCOA2 and fusion with PAX3 (PAX3-NCOA2) in cases 4 and 5 (Fig. 3).
Figure 3.
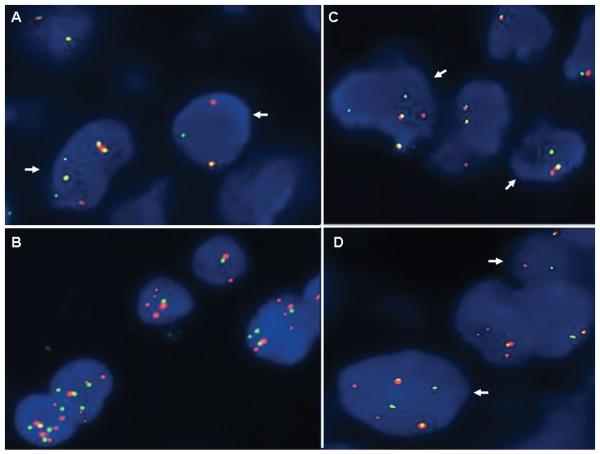
Representative FISH analysis of t(2;2)(p23;q35) and t(2;8)(q35;q12) translocations. A. FISH analysis of Case 1 with the custom designed NCOA1 break-apart probe set demonstrates split orange and green signals (arrows) indicative of a rearrangement of this locus. B. FISH analysis of Case 1 with the PAX3 spanning probe set in orange and the NCOA1 spanning probe set in green demonstrates the presence of juxtaposed or fused orange and green signals consistent with the RT-PCR findings of a PAX3-NCOA1 fusion transcript in this case. C and D. FISH analyses of Case 4 with the custom designed PAX3 and NCOA2 break-apart probe sets, respectively, demonstrate split orange and green signals (arrows) indicative of a rearrangement of each of these loci.
Characterization of the PAX3-NCOA1 and PAX3-NCOA2 Translocations
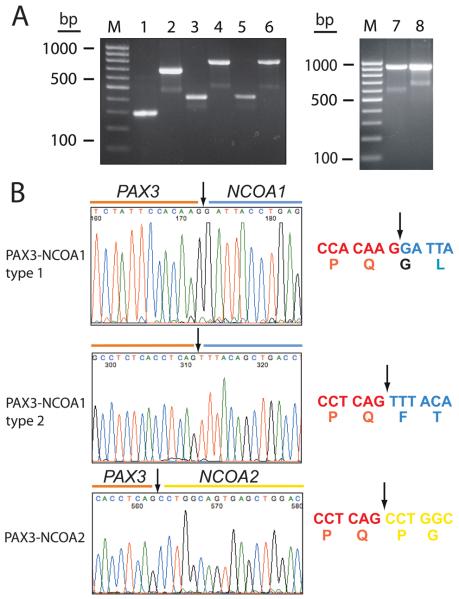
Sequence analysis of the PAX3 3′-RACE PCR products of case 2 revealed fusion of the first 7 exons of PAX3 to the last 10 exons of the nuclear receptor coactivator gene NCOA1. Subsequent RT-PCR analysis with PAX3 forward and NCOA1 reverse primers of this case and case 3 showed that the fusion cDNA junctions for PAX3-NCOA1 were identical (fusion of exon 7 of PAX3 with exon 11 of NCOA1) (Figs. 4 and 5). In contrast, RT-PCR analysis of the ARMS specimen of case 1 revealed a gel electrophoresis amplification product that was slightly smaller in size than the products of cases 2 and 3 (Fig. 4). Consequent sequence analysis of this smaller product demonstrated an in-frame fusion of PAX3 exon 6 with NCOA1 exon 12, a PAX3-NCOA1 fusion transcript similar to that described by Wachtel et al., (2004) (Figs. 4 and 5).
Figure 4.
Representative RT-PCR and sequence analyses for chimeric transcripts in Cases 1-5. A. Detection of PAX3-NCOA1 transcripts in Case 1 (lanes 1 and 2); 2 (lanes 3 and 4) and 3 (lanes 5 and 6). Primers used for lanes 1, 3 and 5 were PAX3-32 and NCOA1-33; for lanes 2, 4 and 6, PAX3-34 and NCOA1-35. Detection of PAX3-NCOA2 transcripts in Cases 4 (lane 7) and 5 (lane 8). Primers used were PAX3-41 and NCOA2-48. B. Sequence alignment of the PAX3-NCOA1 and PAX3-NCOA2 breakpoint regions. Arrows depict the fusion point. Single letter amino acid code is displayed beneath the nucleotide sequence.
Figure 5.
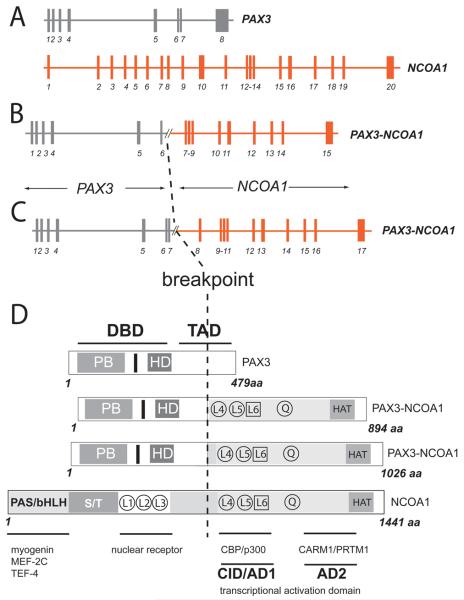
Comparison of wild type and fusion products associated with the t(2;2)(p23;q35) translocation. A-C. Genomic structure of PAX3, NCOA1, PAX3-NCOA1 fusion type 1 (PAX3 exons 1-6 and NCOA1 exons 12-20), and PAX3-NCOA1 type 2 fusion (PAX3 exons 1-7 and NCOA1 exons 11-20), D. Structure of the proteins involved in the fusion. Interacting proteins are displayed as bars. The letters within the bars designate conserved domains (PB, paired domain; HD, homeodomain of the PAX3 protein; and, bHLH/PAS, receptor nuclear translocator domain, involved in DNA binding). S/T represents the serine-threonine-rich region. Transcriptional domains of PAX3 are DBD, DNA binding domain and TAD, transcriptional activation domain. The two transcriptional activator domains of NCOA1 are CID/AD1 and AD2. Encircled L4, and L5 are atypical LXXLL a-helix motifs and boxed L6 is an atypical LXXLL motif, HAT indicates the histone acetyltransferase domain. Factors that interact with specific functional domains are listed beneath the lines of the corresponding domain bars.
The PAX3-NCOA1 fusion protein of case 1 is composed of the DNA-binding domains of PAX3 (paired-box and homeodomain; 319 N-terminal AA) and the C-terminal region of NCOA1 presumably functioning as a transactivation domain (includes the CID/AD1 domain, the Q-rich region and the AD2 domain covering 575 AA), (Fig. 5). The resulting chimeric protein (1,026 residues in total) for cases 2 and 3 includes the initial 391 AA of PAX3 joined to the C-terminal 635 AA of NCOA1 (Fig. 5). Both DNA-binding domains of PAX3 (paired-box and homeodomain) and the interaction domain (CID/AD1), the Q-rich region and the transactivation domain 2 (AD2) of NCOA1 are incorporated into the fusion protein (Fig. 5). We refer to the PAX3-NCOA1 chimeric protein in case 1 as a type 1 fusion and the chimeric proteins of cases 2 and 3 as a type 2 fusion.
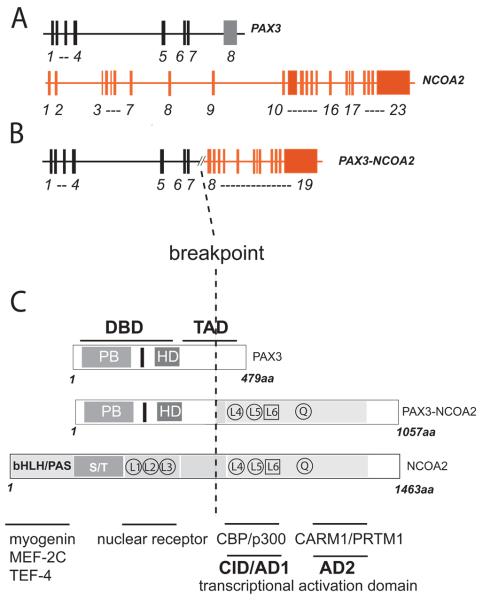
Cloning and sequencing of the RACE-PCR products of case 4 revealed that exon 7 of PAX3 was fused to exon 12 of the NCOA2 gene, the second member of the nuclear receptor transcriptional coactivator family of genes (Figs. 4 and 6). Subsequent RT-PCR analysis with PAX3 forward and NCOA2 reverse primers of this case and case 5 which also exhibited a 2;8 translocation karyotypically, showed identical PAX3-NCOA2 fusion breakpoints (Fig. 6). Reciprocal NCOA2-PAX3 transcripts were not detected. The full-length cDNA construct extended 4,724 nucleotides of which 3,171 are coding. The resulting PAX3-NCOA2 fusion protein is 1057 AA long and consists of the 391 N-terminal AA of PAX3 (DNA-binding domains) and the 666 C-terminal AA of NCOA2 (CID/AD1 domain, the Q-rich region and the AD2 domain 2), (Fig. 6).
Figure 6.
A and B. Genomic structure of PAX3, NCOA2 and the PAX3-NCOA2 fusion (PAX3 exons 1-7 and NCOA2 exons 12-23). C. PAX3, NCOA2 and PAX3-NCOA2 proteins. Refer to Figure 4D legend for structure description.
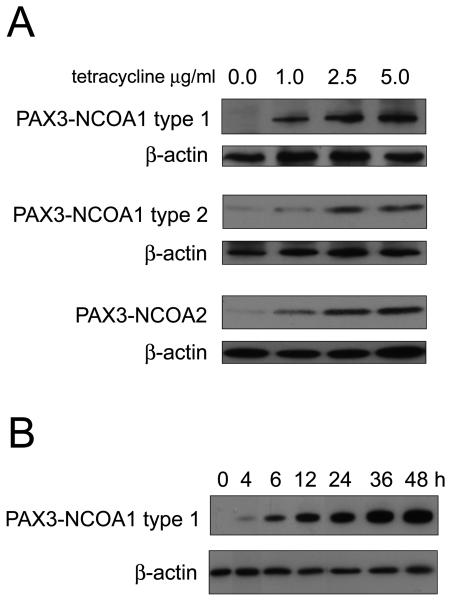
In an effort to examine the oncogenic consequences of PAX3-NCOA1 type 1, PAX3-NCOA1 type 2, and PAX3-NCOA2 expression, a cell culture system using a tetracycline regulated mammalian expression system (T-REx™ system, Invitrogen) was employed. In this model system, NIH 3T3 cells were transfected with pcDNA6/TR plasmid expressing the tet repressor. Following transfection several clones expressing the tet repressor were selected. NIH 3T3-19.1 was preferred due its high inducibility by addition of tetracycline and low leakage in the absence of the drug and was used for the subsequent experiments. PAX3-NCOA1 type 1, PAX3-NCOA1 type 2, and PAX3-NCOA2 fusion transcripts were cloned into pcDNA4/TO vector and transfected into NIH-3T3-19.1 cells. After transduction into NIH-3T3-19.1 cells and selection in zeocin, Western blotting demonstrated that the expression level of PAX3-NCOA1 type 1, PAX3-NCOA1 type 2, and PAX3-NCOA2 chimeric genes was regulated by the concentration of tetracycline in the medium (Fig. 7A). Western blotting analysis revealed a rapid induction of PAX3-NCOA1 type 1 proteins as early as 4 hours and reached the peak level at 36 hours (Fig. 7B).
Figure 7.
Tetracycline inducible expression of PAX3-NCOA1 and PAX3-NCOA2 genes in transfected NIH 3T3 cells. A. Representative Western blots demonstrate the induction of varying expression levels of PAX3-NCOA1 type 1, PAX3-NCOA1 type 2 and PAX3-NCOA2 chimeric genes that correspond with different doses of tetracycline. B. Western blot demonstrating the rapid induction of PAX3-NCOA1 type 1 expression in the presence of 2.5 μg/ml tetracycline (cells were harvested at the time points indicated).
Transforming Properties of PAX3-NCOA1 and PAX3-NCOA2
Zeocin resistant polyclonal cells from PAX3-NCOA1 type 1, PAX3-NCOA1 type 2 and PAX3-NCOA2 transfectants were assayed in the presence of 1 μg/ml tetracycline for anchorage independent growth in liquid medium and colony formation in soft agar (Fig. 8). Cells expressing NCOA1 or NCOA2 showed anchorage independent growth (Fig. 8A) and displayed macroscopically visible colonies in soft agar (Fig. 8B). In contrast cells transfected with the empty vector showed no visible colony formation and displayed a flat morphology when grown on plastic (Fig. 8A).
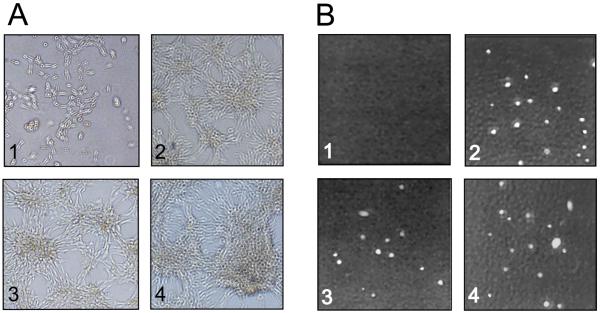
Figure 8.
Effect of PAX3-NCOA1 type 1, PAX3-NCOA1 type 2 and PAX3-NCOA2 expression on NIH3T3 cell growth. A. NIH3T3-Tet-19.1 cells transfected with plasmids pcDNA4/TO (1), PAX3-NCOA1-type 1 (2), PAX3-NCOA1-type 2 (3) and PAX3-NCOA2 (4) in pcDNA4/TO and grown in the presence of 1 μg/ml tetracycline. B. Representative agar assay images acquired 2 weeks after cells were plated at low density and grown in the presence of 1 μg/ml tetracycline show formation of macroscopic colonies in the PAX3-NCOA1-type 1 (2), PAX3-NCOA1-type 2 (3) and PAX3-NCOA2 (4) plates but not in the pcDNA4/TO (1) plate.
The CID/AD1 domain of NCOA proteins is important to recruit CBP/p300 into the nuclear receptor mediated transcriptional complex to acetylate histone and non-histone proteins and to promote transcription (Voegel et al., 1996; Li et al., 1997; Onate et al., 1998). The second transcriptional activation domain (AD2), also located at the C terminus of NCOA proteins, is responsible for interaction with histone methyltransferases, coactivator-associated arginine methyltransferase-1 and protein arginine methyltransferase 1 (Chen et al., 1999; Koh et al., 2001). Both CID/AD1 and AD2 are important functional domains and both are retained in the PAX3-NCOA1 and PAX3-NCOA2 fusion proteins.
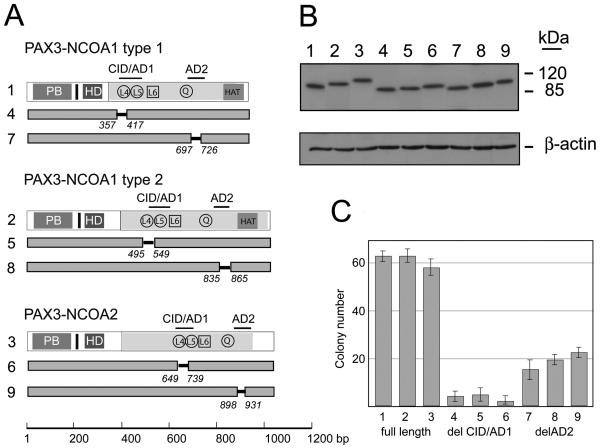
To explore the involvement of the CID/AD1 and AD2 domains in the oncogenic activity of PAX3-NCOA1 and PAX3-NCOA2, targeted deletions were introduced. The AD1 and AD2 deletions represent in-frame deletions from Ser357 to Gly411(CID, AD1 domain) and from ser697 to Pro727 (AD2 domain) of the PAX3-NCOA1 type 1 protein. Corresponding deletions in PAX3-NCOA1 type 2 proteins were from Ser495 to Gly549 (AD1 /CID domain) and from Ser835 to Pro865 (AD2 domain) (Fig. 9A). The in-frame deletions in PAX3-NCOA2 were between Phe649 and Gly739 (CID/AD1 domain) and Val898 and Met931 (AD2 domain). Western blotting analysis verified the sizes and expression of the mutated proteins in NIH 3T3-19.1 cells (Fig. 9B).
Figure 9.
Analyses of transforming activities of PAX3-NCOA1 and PAX3-NCOA2 fusion proteins and deletion mutants. A. Schematic structure of PAX3-NCOA1, PAX3-NCOA2 and deletion mutants. Solid bars with amino acid position displays the regions deleted. B. Immunoblot analyses of PAX3-NCOA1, PAX3-NCOA2 and deletion mutants in NIH3T3-Tet-19.1.E. C. Soft agar colony assay of pcDNA4/TO vector (1), pcDNA4/TO-PAX3-NCOA1 type 1, pcDNA4/TO-PAX3-NCOA1 type 2, and pcDNA4/TO-PAX3-NCOA2 transduced NIH3T3-Tet-19.1 cells. Cells were incubated in the presence of 1μg/ml tetracycline and scored after 14 days. Three plates were counted for each construct.
NIH 3T3-Tet-19.1 cells transduced with the PAX3-NCOA1 or PAX3-NCOA2 lacking the AD2 domain result in decreased number of soft agar colonies (Fig. 9C). The deletion of CID/AD1 domain showed dramatic decrease in their ability to form colonies in soft agar suggesting (Fig. 9C) that the integrity of this region is important in oncogenic activity of the chimeric protein.
DISCUSSION
At least 70% of alveolar rhabdomyosarcomas are characterized by the expression of one of two chimeric proteins generated by translocations that fuse the FOXO1 gene with the PAX3 or PAX7 gene; the remainder of cases lack these translocations (fusion transcript negative ARMS). Following the design of novel PAX3, PAX7 and FOXO1 FISH probe sets, we uncovered a PAX3 rearrangement in an ARMS case negative for PAX3-FOXO1 and PAX7-FOXO1 fusion transcripts (Nishio et al., 2006). This finding, suggesting the presence of a PAX3 variant translocation, prompted an investigation of the potential translocation gene partner in this case and three additional fusion transcript negative ARMSs and one congenital ERMS, each also suspicious for containing PAX3 variant translocations based on conventional cytogenetic and/or PAX3 FISH analysis. Newly recurrent chimeric genes, as well as novel ones characterized by fusion of the PAX3 gene with either NCOA1 or NCOA2 were identified in each case. PAX3-NCOA1 is structurally heterogeneous and two types of in-frame PAX3-NCOA1 chimeric transcripts were discovered. We have defined a “type 1” PAX3-NCOA1 fusion transcript as one with the fusion breakpoint located in intron 6 of PAX3 and in intron 12 of NCOA1; a rearrangement that has also been observed in an isolated case reported by Wachtel et al. (2004) among a series of 29 RMSs subjected to gene expression profiling. The designation “type 2” PAX3-NCOA1 chimeric transcript is proposed for fusion of PAX3 exon 7 with NCOA1 exon 11.
The clinicohistopathologic findings of Case 5 were consistent with the diagnosis of congenital ERMS (Meloni-Ehrig et al., 2009). In contrast to ARMS, ERMS has not been classified as a translocation sarcoma. Rather, ERMSs frequently exhibit a recurrent pattern of chromosomal imbalances (gain of all or portions of chromosomes 2, 7, 8, 11, 12, 13 and/or 20, with or without loss of 22) (Polito et al., 1999; Bridge et al., 2000). To the best of our knowledge, this is the third reported case of ERMS with cytogenetic abnormalities involving chromosomal breakpoints 2q35 and 8q13 (Hayashi et al., 1988; Yoshino et al., 2005). Recently, Davicioni et al. (2009) described a genomic-based classification scheme that is at variance with conventional histopathological schemes. Specifically, the gene expression profiles and LOH patterns of PAX-FOXO1 negative ARMSs were indistinguishable from conventional ERMSs and thus, the authors concluded that RMS can be effectively divided into two main molecular classes (PAX-FOXO1 positive and negative RMS). Moreover, it was proposed that it may be more appropriate to define tumors both by histological appearance (alveolar, embryonal, and the embryonal variants) and by fusion status (PAX-FOXO1 positive and negative). This is an interesting concept as it relates to the presence of PAX3 variant translocations, a genotype historically linked with an alveolar phenotype, in cases of histologic ERMS.
In contrast to FOXO1, NCOA1 and NCOA2 do not encode for transcriptional factors. NCOA1 and NCOA2, in addition to NCOA3 (20q13.12), are members of the p160 nuclear receptor transcriptional coactivator family. The proteins encoded by these genes have an overall sequence similarity of 50–55% between the three members (Xu and O'Malley, 2002; Xu and Li, 2003). Their most conserved N-terminal bHLH/PAS receptor nuclear translocator domain is involved in DNA binding and heterodimerization between proteins containing the same motifs. The intrinsic transcriptional activation domain, CID/AD1 is responsible for recruiting acetyltransferases including CBP/p300 and p/CAF for chromatin remodeling (Voegel et al., 1996; Li et al., 1997; Onate et al., 1998). The second transcriptional activation domain (AD2) is located at the C-terminus and is responsible for the interaction with the histone methyltransferases CARM1 and PRMT1 (Chen et al., 1999; Koh et al., 2001). In addition, NCOA1 and NCOA3 possess weak intrinsic HAT activities at the C-terminal region (Chen et al., 1997; Spencer et al., 1997). Moreover, NCOA proteins contain specific LXXLL motifs (where L is leucine and X is any amino acid), which arrange the interaction with nuclear receptors via the NID (three motifs), as well as with p300/CBP through the CID/AD1 (two motifs) (Heery et al., 1997; Kalkhoven et al., 1998). Importantly, the NCOA proteins are capable of interacting with multiple nuclear receptors in a ligand-dependent mode and significantly augment nuclear receptor-dependent transcriptional activity (Leo et al., 2000; McKenna and O'Malley, 2002; Xu and Li, 2003).
The structure and function of PAX3 have been well reviewed (Buckingham and Relaix, 2007; Mercado and Barr, 2007). This member of the family of paired box transcription factors contains an N-terminal DNA-binding domain, a complete homeodomain and a proline- serine- and threonine-rich transcriptional activation domain. The PAX3-NCOA1 and PAX3-NCOA2 fusion genes produce mRNAs containing the 5′ end of PAX3 appended in translational frame to the 3′ end of NCOA1 or NCOA2. Similar to PAX-FOXO1 fusion products, PAX3-NCOA fusion products retain the DNA binding domain of PAX3. The CBP/p300 interaction domain, the Q-rich region and AD2 transcriptional activation domain of the nuclear receptor coactivator proteins are also retained within the PAX3-NCOA chimeric products. In addition, the transactivation capability of the PAX3-NCOA1 fusion protein using a luciferase reporter gene system appears to be comparable to PAX3-FOXO1 (Wachtel et al., 2004).
In contrast, the PAX3-NCOA fusions do not retain the PAS/bNLH domain of the nuclear receptor coactivator protein, believed to be involved in DNA-binding and protein heterodimerization, or the receptor interacting domain (RID), which mediates the binding of transcriptional coactivators to nuclear receptors via conserved LXXLL/LXXLL motifs. Therefore, it is unlikely that PAX3-NCOA1 or -NCOA2 fusion proteins are able to interact with any of the upstream components that normally require NCOA1 or NCOA2 as a transcriptional intermediary. Instead, the activity of AD1/CID may directly modulate or augment the transcriptional activity of genes normally regulated by PAX3 through recruiting CBP/p300. Alternatively, the intrinsic HAT and histone methyltransferase activities of the nuclear receptor coactivator moiety of the fusion oncogenes are relevant to putative sarcomagenic alterations in gene expression.
NCOA2 and NCOA3 have been identified in translocation events in other neoplastic entities. The C-terminal portion of NCOA2 retained in the MYST3-NCOA2 fusion protein generated by an inv(8)(p11q13) in acute myelogenous leukemia (AML) (Murati et al., 2004; Carapeti et al., 1998; Liang et al., 1998) and in the ETV6-NCOA2 chimeric protein created by the t(8;12)(q13;p13) in pediatric acute lymphoblastic leukemia (Strehl et al., 2008) is identical to that preserved in the PAX3-NCOA2 fusion. Fusion of the C-terminal portion of NCOA3 to MYST3 has also been observed in an M5-AML (Esteyries et al., 2008). The involvement of the same functional domains of the nuclear receptor coactivators in various oncogenic fusion proteins emphasizes the importance of the CID/AD1 and AD2 regions in tumorigenesis. Fusion between NCOA1 and genes other than PAX3 has not been observed yet.
As demonstrated, a deletion in the CID/AD1 domain of NCOA1 or NCOA2 abrogated soft agar cloning. These data indicate that the conserved LXXLL motifs in CID/AD1 of NCOA1 and NCOA2 play a pivotal role in PAX3-NCOA1 or PAX3-NCOA2 mediated transformation. The activity of NCOA1 and NCOA2 is stimulated through recruiting CBP/p300 (Kim et al., 2001; Wang et al., 2001). CBP/p300 are universal transcription coactivators with HAT activity (Bannister and Kouzarides, 1996; Ogryzko et al., 1996;) that participate in multiple transcriptional events through the regulation of histone acetylation and interaction with the basal transcriptional machinery (Avantaggiati et al., 1997; von Mikecz et al., 2000;). CBP/p300 also contributes to the acetylation of non-histone proteins, such as RB1, E2F, and TP53 and regulates cell growth and differentiation. In the PAX3-NCOA1 and PAX3-NCOA2 fusions, CID, a CBP interacting domain in the nuclear receptor coactivator portion, is retained. This domain is required for NIH3T3 cells to form colonies in soft agar. In contrast, the deletion of AD2 from NCOA1 and NCOA2 produced only a fifty percent reduction in the number of soft agar colonies.
In conclusion, these data suggest that the nuclear receptor coactivator portion of the PAX3-NCOA1 or -NCOA2 fusion proteins is necessary to activate the oncogenic potential of PAX3 by providing activation domains for stimulating transcription or HAT activity for remodeling chromatin. The PAX3 DNA-binding domains could deliver the CBP interaction domain (AD1) and histone methylatransferase domain (AD2) to unique chromosomal locations modifying transcription by allowing binding of CBP or other proteins and/or through remodeling the chromatin around the PAX3 binding site. Additional studies will be needed to determine if the downstream genes and pathways activated by PAX3-NCOA fusions are shared or contrast with those stimulated by PAX-FOXO1 fusions. The discovery of these recurrent, novel translocations and associated fusion oncoproteins should shed light on the relationship between the control of chromatin condensation and gene expression in RMS and may further contribute to directed subtype-specific therapeutics.
Acknowledgments
The authors would like to thank Dr. Beat Schäfer (University Children's Hospital, Zurich, Switzerland) for kindly providing a PAX3-NCOA1 positive RNA control sample. The authors would also like to thank Dr. Gregorio Chejfec (University of Illinois Medical Center, Chicago, IL) and Dr. Diana Corao (DuPont Hospital for Children, Wilmington, DE) for their case material contributions.
Supported by: This work was supported in part by the following: NIH U-10-CA98543-01, NIH RO1-CA89461, UNMC Eppley Pediatric Cancer Award, The Ohio division of the American Cancer Society, and the La Fondation des Gouverneurs de l'Espoir.
References
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Barr FG, Qualman SJ, Macris MH, Melnyk N, Lawlor ER, Strzelecki DM, Triche TJ, Bridge JA, Sorensen PH. Genetic heterogeneity in the alveolar rhabdomyosarcoma subset without typical gene fusions. Cancer Res. 2002;62:4704–4710. [PubMed] [Google Scholar]
- Bridge JA, Liu J, Weibolt V, Baker KS, Perry D, Kruger R, Qualman S, Barr F, Sorensen P, Triche T, Suijkerbuijk R. Novel genomic imbalances in embryonal rhabdomyosarcoma revealed by comparative genomic hybridization and fluorescence in situ hybridization: an intergroup rhabdomyosarcoma study. Genes Chromosomes Cancer. 2000;27:337–344. doi: 10.1002/(sici)1098-2264(200004)27:4<337::aid-gcc1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annual Rev Cell Dev Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- Carapeti M, Aguiar RC, Goldman JM, Cross NC. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91:3127–3133. [PubMed] [Google Scholar]
- Carr PA, Park JS, Lee YJ, Yu T, Zhang S, Jacobson JM. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Davicioni E, Anderson MJ, Finckenstein FG, Lynch JC, Qualman SJ, Shimada H, Schofield DE, Buckley JD, Meyer WH, Sorensen PH, Triche TJ. Molecular classification of rhabdomyosarcoma--genotypic and phenotypic determinants of diagnosis: a report from the Children's Oncology Group. Am J Pathol. 2009;174:550–564. doi: 10.2353/ajpath.2009.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, D'Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- Deguchi K, Ayton PM, Carapeti M, Kutok JL, Snyder CS, Williams IR, Cross NC, Glass CK, Cleary ML, Gilliland DG. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3:259–271. doi: 10.1016/s1535-6108(03)00051-5. [DOI] [PubMed] [Google Scholar]
- Esteyries S, Perot C, Adelaide J, Imbert M, Lagarde A, Pautas C, Olschwang S, Birnbaum D, Chaffanet M, Mozziconacci MJ. NCOA3, a new fusion partner for MOZ/MYST3 in M5 acute myeloid leukemia. Leukemia. 2008;22:663–665. doi: 10.1038/sj.leu.2404930. [DOI] [PubMed] [Google Scholar]
- Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, Emanuel BS, Rovera G, Barr FG. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;3:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Inaba T, Hanada R, Yamamoto K. Translocation 2;8 in a congenital rhabdomyosarcoma. Cancer Genet Cytogenet. 1988;30:343–345. doi: 10.1016/0165-4608(88)90208-7. [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Kalkhoven E, Valentine JE, Heery DM, Parker MG. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapels KM, Nishio J, Zhou M, Qualman SJ, Bridge JA. Embryonal rhabdomyosarcoma with a der(16)t(1;16) translocation. Cancer Genet Cytogenet. 2007;174:68–73. doi: 10.1016/j.cancergencyto.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Kim MY, Hsiao SJ, Kraus WL. A role for coactivators and histone acetylation in estrogen receptor alpha-mediated transcription initiation. EMBO J. 2001;20:6084–6094. doi: 10.1093/emboj/20.21.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh SS, Chen D, Lee YH, Stallcup MR. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J Biol Chem. 2001;276:1089–1098. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- Li S, Aufiero B, Schiltz RL, Walsh MJ. Regulation of the homeodomain CCAAT displacement/cut protein function by histone acetyltransferases p300/CREB-binding protein (CBP)-associated factor and CBP. Proc Natl Acad Sci USA. 2000;97:7166–7171. doi: 10.1073/pnas.130028697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Prouty L, Williams BJ, Dayton MA, Blanchard KL. Acute mixed lineage leukemia with an inv(8)(p11q13) resulting in fusion of the genes for MOZ and TIF2. Blood. 1998;92:2118–2122. [PubMed] [Google Scholar]
- Lugo TG, Witte ON. The BCR-ABL oncogene transforms Rat-1 cells and cooperates with v-myc. Mol Cell Biol. 1989;9:1263–1270. doi: 10.1128/mcb.9.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW. Minireview: nuclear receptor coactivators, an update. Endocrinology. 2002;143:2461–2465. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- Meloni-Ehrig A, Smith B, Zgoda J, Greenberg J, Perdahl-Wallace E, Zaman S, Mowrey P. Translocation (2;8)(q35;q13): a recurrent abnormality in congenital embryonal rhabdomyosarcoma. Cancer Genet Cytogenet. 2009;191:43–45. doi: 10.1016/j.cancergencyto.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Mercado GE, Barr FG. Fusions involving PAX and FOX genes in the molecular pathogenesis of alveolar rhabdomyosarcoma: recent advances. Curr Mol Med. 2007;7:47–61. doi: 10.2174/156652407779940440. [DOI] [PubMed] [Google Scholar]
- von Mikecz A, Zhang S, Montminy M, Tan EM, Hemmerich P. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J Cell Biol. 2000;150:265–273. doi: 10.1083/jcb.150.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murati A, Adélaïde J, Mozziconacci MJ, Popovici C, Carbuccia N, Letessier A, Birg F, Birnbaum D, Chaffanet M. Variant MYST4-CBP gene fusion in a t(10;16) acute myeloid leukaemia. Br J Haematol. 2004;125:601–604. doi: 10.1111/j.1365-2141.2004.04960.x. [DOI] [PubMed] [Google Scholar]
- Nishio J, Althof PA, Bailey JM, Zhou M, Neff JR, Barr FG, Parham DM, Teot L, Qualman SJ, Bridge JA. Use of a novel FISH assay on paraffin-embedded tissues as an adjunct to diagnosis of alveolar rhabdomyosarcoma. Lab Invest. 2006;86:547–556. doi: 10.1038/labinvest.3700416. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Onate SA, Boonyaratanakornkit V, Spencer TE, Tsai SY, Tsai MJ, Edwards DP, O'Malley BW. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- Parham DM, Ellison DA. Rhabdomyosarcomas in adults and children: an update. Arch Pathol Lab Med. 2006;130:1454–1465. doi: 10.5858/2006-130-1454-RIAACA. [DOI] [PubMed] [Google Scholar]
- Polito P, Dal Cin P, Sciot R, Brock P, Van Eykan P, Van Den Berghe H. Embryonal rhabdomyosarcoma with only numerical chromosome changes. Case report and review of the literature. Cancer Genet Cytogenet. 1999;109:161–165. doi: 10.1016/s0165-4608(98)00168-x. [DOI] [PubMed] [Google Scholar]
- Qualman SJ, Coffin CM, Newton WA, Hojo H, Triche TJ, Parham DM, Crist WM. Intergroup Rhabdomyosarcoma study: Update for pathologists. Pediatr Dev Pathol. 1998;1:550–561. doi: 10.1007/s100249900076. [DOI] [PubMed] [Google Scholar]
- Raney RB, Anderson JR, Barr FG, Donaldson SS, Pappo AS, Qualman SJ, Wiener ES, Maurer HM, Crist WM. Rhabdomyosarcoma and undifferentiated sarcoma in the first two decades of life: a selective review of intergroup rhabdomyosarcoma study group experience and rationale for Intergroup Rhabdomyosarcoma Study V. J Pediatr Hematol Oncol. 2001;23:215–220. doi: 10.1097/00043426-200105000-00008. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Slovak ML, Campbell LJ, editors. An International System for Human Cytogenetic Nomenclature Basel. Karger, ISCN; 2009. [Google Scholar]
- Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, Bridge JA, Crist WM, Triche TJ, Barr FG. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J Clin Oncol. 2002;11:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- Strehl S, Nebral K, König M, Harbott J, Strobl H, Ratei R, Struski S, Bielorai B, Lessard M, Zimmermann M, Haas OA, Izraeli S. ETV6-NCOA2: a novel fusion gene in acute leukemia associated with coexpression of T-lymphoid and myeloid markers and frequent NOTCH1 mutations. Clin Cancer Res. 2008;14:977–983. doi: 10.1158/1078-0432.CCR-07-4022. [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- Wachtel M, Dettling M, Koscielniak E, Stegmaier S, Treuner J, Simon-Klingenstein K, Bühlmann P, Niggli FK, Schäfer BW. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64:5539–5545. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem. 2001;276:18375–18383. doi: 10.1074/jbc.M100800200. [DOI] [PubMed] [Google Scholar]
- Yoshino K, Takeuchi M, Nakayama M, Suehara N. Congenital cervical rhabdomyosarcoma arising in one fetus of a twin pregnancy. Fetal Diagn Ther. 2005;20:291–295. doi: 10.1159/000085088. [DOI] [PubMed] [Google Scholar]
- Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17:1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- Xu J, O'Malley BW. Molecular mechanisms and cellular biology of the steroid receptor coactivator (SRC) family in steroid receptor function. Rev Endocr Metab Disord. 2002;3:185–192. doi: 10.1023/a:1020016208071. [DOI] [PubMed] [Google Scholar]
- Zhuravleva J, Paggetti J, Martin L, Hammann A, Solary E, Bastie JN, Delva L. MOZ/TIF2-induced acute myeloid leukaemia in transgenic fish. Br J Haematol. 2008;143:378–382. doi: 10.1111/j.1365-2141.2008.07362.x. [DOI] [PubMed] [Google Scholar]