Abstract
Columns containing immobilized lipoproteins were prepared for the analysis of drug interactions with these particles by high-performance affinity chromatography. This approach was evaluated by using it to examine the binding of high density lipoprotein (HDL) to the drugs propranolol or verapamil. HDL was immobilized by the Schiff base method onto silica and gave HPLC columns with reproducible binding to propranolol over four to five days of continuous operation at pH 7.4. Frontal analysis experiments indicated that two types of interactions were occurring between R/S-propranolol and HDL at 37°C: saturable binding with an association equilibrium constant (Ka) of 1.1–1.9 × 105 M−1, and non-saturable binding with an overall affinity constant (n Ka) of 3.7–4.1 × 104 M−1. Similar results were found at 4 and 27°C. Verapamil also gave similar behavior, with a Ka of 6.0 × 104 M−1 at 37°C for the saturable sites and a n Ka value for the non-saturable sites of 2.5 × 104 M−1. These measured affinities gave good agreement with solution-phase values. The results indicated HPAC can be used to study drug interactions with HDL, providing information that should be valuable in obtaining a better description of how drugs are transported within the body.
Keywords: high density lipoprotein, propranolol, verapamil, high-performance affinity chromatography, drug binding, frontal analysis
INTRODUCTION
The binding of drugs with proteins, cell receptors, enzymes, immunoglobins and other biological agents is important in determining the activity, pharmacokinetics and toxicity of these drugs in the human body [1–3]. Many drugs can also undergo reversible interactions with serum proteins and lipoproteins in blood. These interactions typically have association equilibrium constants or overall affinities in the range of 103 to 106 M−1 and can play an important role in affecting the distribution, delivery, metabolism, and excretion of drugs [1–6]. As a result, the pharmaceutical industry often performs binding studies with these agents when designing a new drug and when determining which mode of drug delivery might treat a disease most efficiently [3,4,7].
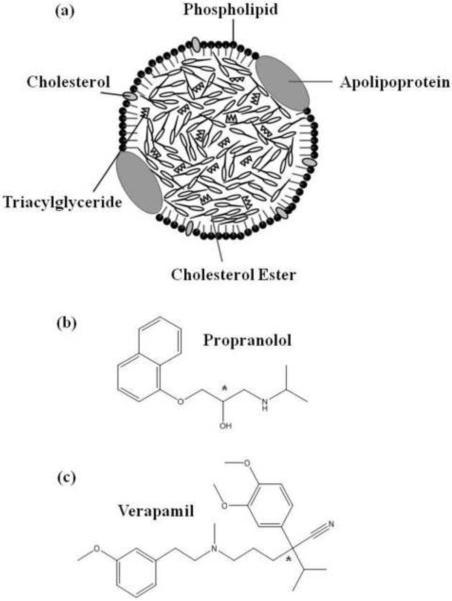
As shown in Figure 1(a), a lipoprotein is a soluble macromolecular complex of proteins and lipids that transports hydrophobic compounds such as cholesterol and triglycerides [7–9]. In humans, lipoproteins are classified according to their density, giving categories that include low density lipoprotein (LDL) and high density lipoprotein (HDL) [7,9,11–15]. In addition to binding cholesterol and triglycerides, these lipoproteins are also known to bind several basic or neutral drugs in blood [10].
Figure 1.
(a) General structure of high density lipoprotein (HDL) and other lipoprotein particles, and the structures of (b) propranolol and (c) verapamil, in which the asterisks indicate the locations of the chiral centers in these drugs.
Two drugs known to bind with HDL are propranolol and verapamil (see structures in Figure 1(b–c)). Equilibrium dialysis has previously been used to examine the binding of racemic propranolol with HDL [16]. High-performance frontal analysis/capillary electrophoresis has also been utilized to study interactions of propranolol, verapamil and other drugs to HDL or other lipoproteins [17–19]. Table 1 summarizes the binding constants that have been measured for propranolol and verapamil with HDL in these reports [16–18]. These earlier studies have used a model based on a single, non-saturable interaction between HDL and propranolol or verapamil, with estimated overall affinities (nKa) in the range of 104 M−1 [16–18].
Table 1.
Binding parameters previously reported for the interactions of propranolol and verapamil with HDL
| Analyte | Overall affinity, n Ka (M−1)a | Method [Ref.] | Experimental Conditions |
|---|---|---|---|
| Racemic | pH 7.4 Phosphate buffer (0.66 M) | ||
| Propranolol | 1.60 ( ± 0.14) × 104 | Equilibrium dialysis [16] | 13 μM HDL, 37°C 0.001–1000μM Propranolol |
| R-Propranolol | 2.38 ( ± 0.22) × 104 | High-performance Frontal analysis/capillary | pH 7.4 Phosphate buffer (I = 0.17) 14.6 μM HDL, 37°C |
| S-Propranolol | 2.43 ( ± 0.15) × 104 | electrophoresis [17] | 25–150 μM Propranolol |
| R-Verapamil | 2.75 ( ± 0.61) × 104 | High-performance Frontal analysis/capillary | pH 7.4 Phosphate buffer (I = 0.17) 14.6 μM HDL, 37°C |
| S-Verapamil | 2.81 ( ± 0.33) × 104 | electrophoresis [18] | 25–100 μM Verapamil |
In this study, high-performance affinity chromatography (HPAC) will be evaluated and employed as a tool to provide a more detailed analysis of the interactions between HDL and propranolol or verapamil. HPAC has been successfully utilized in the past to examine the binding of drugs with serum proteins such as human serum albumin (HSA) and α1-acid glycoprotein (AGP) [3–6,20–26]. However, no known previous reports have used HPAC to examine drug interactions with lipoproteins. The advantages of using HPAC for such work include the speed, ease of automation, and precision of this method; this technique has also been shown in many drug binding studies with HSA, and more recently with AGP, to give good correlation versus solution-phase reference methods [3,5,6,20,21,24].
In this report, columns containing immobilized HDL will first be prepared and their stability will be examined through zonal elution studies. Binding of the immobilized HDL to propranolol and verapamil will then be investigated by using frontal analysis. The results will be compared to data obtained in previous work with soluble HDL. These experiments should make it possible to determine the feasibility of using immobilized lipoproteins such as HDL in drug binding studies based on HPAC. The results should also provide further information regarding the nature of the interactions between propranolol and verapamil with HDL and give a more complete picture of how these drugs are transported in blood. This information should be valuable in future work as this approach is applied to other drugs and lipoproteins.
EXPERIMENTAL
Reagents
The human HDL (catalog number L1567, lot no. B73112), separate enantiomers of R- and S-propranolol, and racemic R/S-verapamil were purchased from Sigma (St. Louis, MO, USA). Nucleosil Si-300 silica (7 μm particle diameter, 300 Å pore size) was obtained from Macherey Nagel (Düren, Germany). Reagents for the bicinchoninic acid (BCA) protein assay were from Pierce (Rockford, IL, USA). An assay kit for measuring total cholesterol was purchased from Wako (Richmond, VA, USA). All other chemicals were of the highest grades available. All solutions were prepared using water from a Nanopure purification system (Barnstead, Dubuque, IA, USA) that was filtered using Osmonics 0.22 μm nylon filters from Fisher Scientific (Pittsburgh, PA, USA).
Apparatus
This chromatographic system consisted of two PU-980 pumps (Jasco, Tokyo, Japan), one LabPro injection valve (Rohnert Park, FL, USA), and a UV/Vis SpectroMonitor 3200 variable wavelength absorbance detector from LDC Thermoseparations (Riviera Beach, FL, USA). Chromatographic data were collected and processed using programs based on Labview 5.1 or 7.0 (National Instruments, Austin, TX, USA). Support materials were placed into columns using a slurry packer from Alltech (Deerfield, IL, USA). A PolyScience circulating water bath (VWR, Buffalo Grove, IL, USA) was used to control the temperature of the columns and mobile phases.
Preparation of HDL silica
The reactions used to prepare the HDL silica are shown in Figure 2. First, diol-bonded silica was prepared from Nucleosil Si-300 silica by using a previously published procedure (Step 1 in Figure 2) [24,25]. A modified form of the Schiff base method was used to attach HDL to this support [23]. In this method, 0.2 g of diol-bonded silica was placed into 4 ml of a 90:10 (v/v) mixture of acetic acid and water, to which was then added 0.2 g periodic acid (Step 2). This mixture was sonicated under vacuum for 15 min and placed on a wrist action shaker for over 2 h in the dark at room temperature. The resulting aldehyde-activated silica was washed six times with water and four times with pH 6.0, 0.10 M potassium phosphate buffer.
Figure 2.
Immobilization of HDL onto silica by the Schiff base method.
The aldehyde silica was suspended in 1 ml of pH 6.0, 0.10 M potassium phosphate buffer and sonicated for 5 min under vacuum. A 20 mg portion of HDL and 4.3 mg of sodium cyanoborohydride (NaCNBH4) were added to this slurry (Step 3). This mixture was shaken in the dark at 4 °C for 3 days. The resulting immobilized HDL support was washed four times with pH 7.0, 0.10 M potassium phosphate buffer. A 3.4 mg portion of sodium borohydride (NaBH4) was dissolved into 2 ml of pH 7.0, 0.10 M potassium phosphate buffer and added to the HDL support to reduce any remaining aldehyde groups on the silica and to eliminate non-specific biding that might later occur with such groups. This mixture was shaken for 90 min at room temperature. The final HDL support was washed six times using pH 7.0, 0.10 M potassium phosphate buffer. The HDL support was stored in the same pH 7.0 buffer at 4 °C until use. A separate portion of diol silica, as made in Step 1 in Figure 2, was used as the control support in this study.
The protein content of the HDL support was evaluated by using a BCA protein assay [27,28]. An HDL stock solution was prepared in pH 7.4, 0.067 M potassium phosphate buffer and serial dilutions of this stock solution were made to prepare standards for generating a calibration curve. Diol silica was utilized as the blank. After reacting with the BCA reagents, all samples and standard solutions were filtered through a 0.2 μm nylon filter prior to obtaining absorbance readings for their supernatants at 562 nm.
An assay for the total cholesterol content of the HDL silica was performed with a commercial kit based on an enzymatic colorimetric method [28,30]. In this assay, diol silica was again used as the blank. A calibration curve was prepared by using the standard solution provided with the assay kit and diluting this solution with pH 7.4, 0.067 M potassium phosphate buffer. The HDL silica and diol silica control support were suspended in this buffer and an aliquot of a working reagent solution, prepared from the color reagent and buffer solution provided with the assay kit, was added to all samples and standards according to the manufacturer's instructions. After the samples and standards were allowed to react with the reagent, each sample or standard was passed through a 0.22 μm nylon filter before measuring the absorbance of its supernatant at 600 nm.
Chromatographic studies
The HDL silica and diol silica control support were downward slurry packed at 24 MPa (3500 psi) into 1 cm × 2.1 mm i.d. or 5 cm × 2.1 mm i.d. stainless steel columns using pH 7.4, 0.067 M potassium phosphate buffer as the packing solution. The HDL and control columns were stored in pH 7.4, 0.067 M potassium phosphate buffer at 4°C when not in use. Prior to the chromatographic studies, these columns were placed into water jackets from Alltech and connected to a circulating water bath for temperature control (± 0.1°C). All mobile phases were filtered through 0.22 μm nylon filters and degassed under vacuum over 15 min before use on the chromatographic system. The wavelength used to monitor the elution of R- or S-propranolol was 225 nm, and a wavelength of 229 nm was used for verapamil. The elution of sodium nitrate, which was employed as a non-retained marker for the void volume, was monitored at 205 nm.
Zonal elution experiments were conducted to initially evaluate the stability of the HDL columns. The mobile phase was pH 7.4, 0.067 M potassium phosphate buffer that was continuously applied to 1 cm × 2.1 mm i.d. columns containing the HDL silica or diol silica control support. These experiments were conducted at 37 °C over the course of 300 h. A 20 μL sample of 1 μM R- or S-propranolol was injected onto the HDL column or control column at 1.0 ml/min every 12 h to look for any changes in retention. The column void time was measured by injecting 20 μL of 1 μM sodium nitrate. The retention time for each peak was found by using its central moment, as determined by utilizing Peakfit 4.12 (Systat Software, San Jose, CA, USA).
The frontal analysis studies were performed using the 5 cm × 2.1 mm i.d. HDL column and control column. These studies were carried out at 4 °C, 27°C or 37°C and at 1.0 ml/min in the presence of pH 7.4, 0.067 M potassium phosphate buffer. The use of slightly higher or lower flow rates (0.4–1.2 ml/min) gave less than a 2% change in the measured binding capacities. All of these studies were performed within the first 60 h of operation for each new HDL column; this period was well within the usable time range for these columns, as determined in the zonal elution studies (see Results and Discussion). A total of eleven mobile phase solutions containing 0.1–25 μM R- or S-propranolol or six solutions containing 0.25–10 μM R/S-verapamil were applied to the HDL column and control column. The retained analytes were eluted by passing pH 7.4, 0.067 M potassium phosphate buffer through the column prior to the next frontal analysis experiment. The amount of drug required to saturate the HDL column or control column was determined by integration of the resulting breakthrough curve [3] using Labview 5.1.
Interactions with system components other than HDL made up approximately 5–15% of the total retention noted for R- and S-propranolol on an HDL column, and typically 15–22% of the total retention for verapamil on an HDL column (note: such interactions are known to vary from drug-to-drug on a given type of support and should always be evaluated on a case-by-case basis) [3,31]. These interactions with the support did not create any difficulties in this study because this binding was highly reproducible. In addition, an appropriate correction for the void time and this non-specific binding was easily made by subtracting the breakthrough time of the control column from that measured on the HDL column at each concentration of an applied drug, as successfully used in past studies with the same drugs and other HPLC supports or binding agents [5,6,26,32].
RESULTS AND DISCUSSION
General properties of HDL support
The composition of the HDL support was examined by using both a BCA protein assay and a cholesterol assay. Based on the protein assay, the support was found to contain 68 (± 5) mg HDL per gram silica. The cholesterol content of this same support was 3.4 (± 0.4) mg cholesterol per gram silica. During long term storage in pH 7.4, 0.067 M potassium phosphate buffer, the protein and cholesterol content of the HDL support were found to gradually decrease by approximately the same relative amount. In the case of the protein content, there was a decrease of 43 (± 4)% over three months, while the cholesterol content decreased by 56 (± 7)% over the same period of time. However, the relative amount of cholesterol versus protein before and after storage was essentially the same, giving ratios that agreed within ±2 S.D. of their respective values.
It is known that lipoproteins are macromolecular congregates that are held together by non-covalent interactions. As such, these particles could potentially degrade or collapse when used in flow-based system [7,8]. The consistency of the cholesterol-to-protein ratios measured before and after three months of storage for the HDL silica suggests that the loss of either intact or partial lipoprotein particles was responsible for the long term decrease in the support content noted for this material, rather than the more selective loss of only apolipoproteins or cholesterol. The fact that essentially half of the original HDL content still remained after three months of storage in buffer was also encouraging in that it indicated the HDL support should be suitable for use over shorter periods of time in a flow-based system employed in HPAC studies.
Column stability
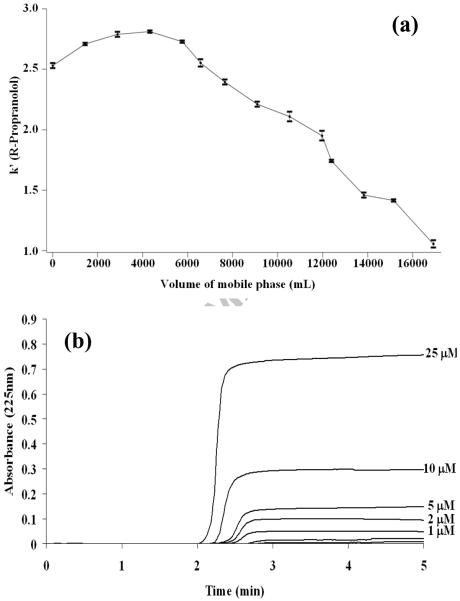
The stability of the HDL support in a flow-based chromatographic system was next examined by using zonal elution experiments in which periodic injections of R- or S-propranolol were made onto this column under controlled temperature and flow rate conditions. Figure 3(a) shows how the retention of R-propranolol changed on the HDL column when samples were injected at 1 mL/min for over the course of 300 h at 37 °C. Similar results were obtained for S-propranolol. Over the first four to five days of continuous use (i.e., 5.8–7.2 L of applied buffer), the HDL column gave reproducible retention with retention times and retention factors that varied by less than ± 5%. The amount of mobile phase applied during four to five days of continuous operation corresponded to over 2 × 105 column volumes for a 1.0 cm × 2.1 mm i.d. column. These data indicated that the HDL column was sufficiently stable for drug binding studies under such conditions.
Figure 3.
(a) Change in the retention factor for R-propranolol as function of mobile phase volume for pH 7.4, 0.067 M potassium phosphate buffer as it was passed through a 1 cm × 2.1 mm i.d. HDL column at 1 ml/min and 37°C. (b) Typical frontal analysis results obtained for the application of R-propranolol to a 5.0 cm × 2.1 mm i.d. HDL column at analyte concentrations of 0.1, 0.2, 0.3, 0.4, 0.5, 1, 2, 5, 10 or 25 μM; these results were also obtained at 1.0 ml/min and 37°C in the presence of pH 7.4, 0.067 M potassium phosphate buffer.
Following the first five days of use, a gradual loss of retention for R- and S-propranolol was noted on the HDL column. After the equivalent of 12 days of continuous operation, the retention factor for R-propranolol had fallen from its original value of 2.5–2.7 to just over 1.0, or a decrease of roughly 9% per day after the first five days. Chiral separations for R- and S-propranolol were not observed on the HDL column at any time during this study, in agreement with previous results reported when using CE to examine the binding of R- and S-propranolol to HDL [17].
Frontal analysis studies of propranolol binding to HDL
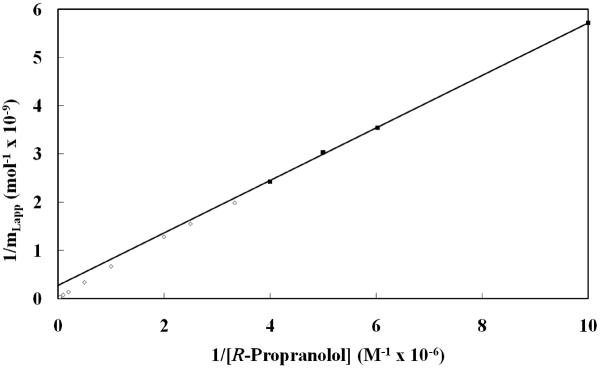
Figure 3(b) shows some typical frontal analysis curves that were obtained for R- or S-propranolol on the HDL columns. These data were all collected within the period of time during which the HDL columns were noted to be stable in the previous section (i.e., within the equivalent of the first four to five days of continuous operation and over the course of approximately 300 applications of sample). The mean position of each breakthrough curve (mLapp) was determined and initially used along with the known concentration of applied drug ([A]) to generate double-reciprocal plots of 1/mLapp versus 1/[A], as shown in Figure 4. This type of plot results in a linear relationship if a single type of binding is occurring; if more than one type of interaction is present, the resulting plot will show negative deviations from a linear response at high analyte concentrations [3]. The plots for R- and S-propranolol at all temperatures examined in this study gave negative deviations from a linear response at large analyte concentrations, or low values of 1/[A]. This behavior indicated that multiple types of interactions were present during the binding of R- and S-propranolol with HDL.
Figure 4.
Double reciprocal plot of frontal analysis data obtained for the binding of R-propranolol to a 5.0 cm × 2.1 mm i.d. HDL column at 37°C in the presence of pH 7.4, 0.067 M potassium phosphate buffer. The linear fit shown was obtained using data points in the upper region of this plot, which are designated by the closed squares (■) and cover R-propranolol concentrations that range from 0.1 μM to 2.5 μM. Data points in the lower region of this plot (i.e., at higher concentrations of R-propranolol) and that showed negative deviations from the linear fit to the upper data points are represented by open squares (□). An expansion of the lower region of the graph is given in the inset. The equation for the best fit line to data represented by the closed squares is y = 5.45 (± 0.07) × 102 x + 2.7 (± 0.6) × 108; the correlation coefficient of this best fit line was 0.99985 (n = 12).
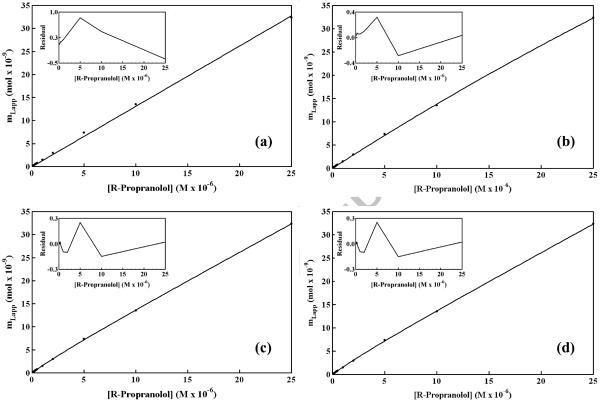
Plots of mLapp versus [A] for R- and S-propranolol were also prepared using the frontal analysis data. Examples of such graphs are given in Figure 5. The non-linear relationship noted in these plots was fit to equations representing four distinct interaction models, as summarized in Table 2. One type of interaction considered was site-specific binding by R- or S-propranolol, as might occur to apolipoproteins on the surface of HDL [16–18]. A second type of interaction that was investigated was non-saturable binding, as might be expected for interactions of R/S-propranolol with the phospholipid layer or interior hydrophobic core of HDL [16–18]. Combinations of these interactions were also considered, such as the presence of two saturable sites or the combination of one saturable site and a group of non-saturable interactions. Table 3 summarizes the best-fit binding parameters that were obtained for each of these models when they were used to examine the frontal analysis data acquired for R- and S-propranolol on the HDL columns.
Figure 5.
Examination of frontal analysis data for R-propranolol on an HDL column at 37°C according to various binding models. These models were as follows: (a) one group of non-saturable interactions, (b) one group of saturable sites, (c) two separate groups of saturable sites, and (d) a group of non-saturable interactions plus a group of saturable sites. The insets show the residual plots for the fit of each model to the experimental data. These results were obtained in the presence of pH 7.4, 0.067 M potassium phosphate buffer. The correlation coefficients were as follows: (a) 0.99937, n=12; (b) 0.99989, n = 12; (c) 0.99996, n = 12; and (d) 0.99996, n = 12. The other best-fit parameters are summarized in Table 3.
Table 2.
Equations for various binding models that were used to fit frontal analysis data obtained for propranolol and verapamil on HDL columns
| Binding model | Predicted response |
|---|---|
| Single non-saturable interaction | mLapp = mL1Ka1[D] |
| Single group of saturable sites | mLapp = (mL1Ka1[D])/(1 + Ka1[D]) |
| Two interactions, saturable site + non-saturable | mLapp = (mL1Ka1[D])/(1 + Ka1[D]) + mL2Ka2[D] |
| Two groups of saturable sites | mLapp = (mL1Ka1[D])/(1 + Ka1[D]) + (mL2Ka2[D])/(1 + Ka2[D]) |
Table 3.
Binding parameters obtained for R-propranolol on an HDL column at 37°C using various binding modelsa
| Binding model | mL1 (mol) | Ka1 (M−1) | mL2 (mol) | Ka2 (M−1) | n Ka (M−1)b |
|---|---|---|---|---|---|
| Single interaction, non-saturable | N/A | N/A | N/A | N/A | 4.4 (± 0.3) × 104 |
| Single groups of saturable sites | 3.0 (± 0.4) × 10−7 | 4.7 (± 0.7) × 103 | N/A | N/A | N/A |
| Two interactions, saturable + non-saturable | 2.2 (± 0.7) × 10−9 | 1.9 (± 0.8) × 105 | N/A | N/A | 4.1 (± 0.7) × 104 |
| Two groups of saturable sites | 2.1 (± 4.2) × 10−9 | 2.0 (± 3.2) × 105 | 8 (± 280) × 10−6 | 1.4 (± 49) × 102 | N/A |
The numbers in parentheses represent a range of ± 1 S.D. All of these results were measured at 37°C in the presence of pH 7.4, 0.067 M potassium phosphate buffer.
The value for n Ka for a non-saturable interaction was obtained by dividing the best-fit result for mL Ka by the estimated moles of HDL in the column. This latter value was obtained by using the measured protein content of the HDL support and an average molar mass for HDL of 1.8 × 105 g/mol [17].
It was already known from double reciprocal plots, such as those given in Figure 4, that multiple interactions were present for R- and S-propranolol with the immobilized HDL. Thus, it was not surprising that the non-reciprocal plots in Figure 5 gave a better fit with models that involved more than one type of interaction. Although the single interaction models did give good correlation coefficients for the plots in Figure 5 (r = 0.9993 to 0.9999), the residual plots for these fits (see insets) represented a non-random distribution of data points about the best-fit lines. In contrast to this, the multiple interaction models also gave large correlation coefficients (i.e., 0.9998 to 0.9999 in all cases) as well as lower residuals and a more random distribution of data about the best-fit lines than seen with the single interaction models.
The model giving the best overall fit to the data was one in which there was a high affinity group of saturable sites and a lower affinity non-saturable interaction. It should be noted that this multiple interaction model gave essentially the same correlation coefficient and residual plot in Figure 5(c) as was obtained for the two-site saturable model in Figure 5(d) (e.g., values for r greater than 0.9999). However, these two models did differ significantly in the precision of their best-fit parameters, as shown in Table 3 by the standard deviations listed for Ka1, mL1 and nKa in the saturable/non-saturable model versus those listed for Ka1, mL1, Ka2 and mL1 in the two-site saturable model. This result can be explained by the fact that the equation for the two-site saturable model approaches the equation for the combined saturable/non-saturable interactions as Ka2 [D] becomes much smaller than one (see expressions in Table 2). This situation was found to occur with the best-fit parameters determined for the two-site saturable model in Table 3 and explains the large uncertainly in these particular parameters for the propranol/HDL system. This observation also explains why the residual plots in Figures 5(c) and Figure 5(d) were so similar because they were actually describing the same model in which there was a high affinity saturable site and a set of lower affinity and essentially non-saturable interactions.
Temperature studies and binding mechanism for propranolol with HDL
Table 4 summarizes the results that were obtained for R- and S-propranolol with HDL at several temperatures when work was continued with a saturable/non-saturable multiple interaction model. For R-propranolol, the saturable interaction had an association equilibrium constant (Ka1) of 1.9 (± 0.8) × 105 at 37°C, which represents relatively high affinity and specific binding for the interaction of a drug with a serum binding agent [2–6]. This binding is probably occurring between R-propranolol and apolipoproteins on HDL, as supported by a comparison of mL1 with the moles of apolipoproteins estimated to be present in the HDL column (see following discussion). The second type of interactions had an overall affinity of 4.1 (± 0.3)×104 M−1 at 37°C, which represents lower affinity and non-specific binding. This second interaction is believed to occur between R-propranolol and phospholipids or the non-polar core of HDL, as has been suggested in previous studies [16,17]. Similar values were found for S-propranolol at 37°C.
Table 4.
Binding parameters determined for propranolol and verapamil on an HDL column using a two site, saturable/one non-saturable binding modela
| Temperature (°C) | Ka1 (M−1) | mL1 (mol) | n Ka2 (M−l)b | |
|---|---|---|---|---|
| R-Propranolol | 4 | 1.4 (± 0.2) × 105 | 6.4 (± 0.8) × 10−9 | 3.7 (± 0.3) × 104 |
| 27 | 1.4 (± 0.2) × 105 | 5.4 (± 0.6) × 10−9 | 3.9 (± 0.3) × 104 | |
| 37 | 1.9 (± 0.8) × 105 | 2.2 (± 0.7) × 10−9 | 4.1 (± 0.3) × 104 | |
| S-Propranolol | 4 | 1.3 (± 0.5) × 105 | 7.8 (± 2.5) × 10−9 | 3.9 (± 0.3) × 104 |
| 27 | 1.6 (± 0.1) × 105 | 3.9 (± 0.2) × 10−9 | 4.0 (± 0.3) × 104 | |
| 37 | 1.1 (± 0.1) × 105 | 4.5 (± 0.2) × 10−9 | 3.7 (± 0.2) × 104 | |
| R/S-Verapamil | 37 | 6.0 (± 2.1) × 104 | 5.4 (± 16) × 10−8 | 2.5 (± 1.5) × 104 |
The numbers in parentheses represent a range of ± 1 S.D. All of these results were measured in the presence of pH 7.4, 0.067 M potassium phosphate buffer.
The value for n Ka2 for the non-saturable interaction was calculated from the best-fit result for mL2 Ka2, as described in Table 3.
The effect of temperature on the interactions between R- and S-propranolol and HDL was also studied. The binding parameters determined for each enantiomer of propranolol with HDL are provided in Table 4. It was found that a change in temperature had no appreciable effect on either the association equilibrium constants or binding capacities that were measured between 4°C and 37°C. The Ka1 values obtained for both R- and S-propranolol were consistently in the range of 1.1 to 1.9×105 M−1 under these temperature conditions and the nKa2 values were all in the range of 3.7 to 4.1×104 M−1. The correlation coefficients were greater than 0.9999 for all frontal analysis plots that were analyzed according to a saturable/non-saturable model. The Ka1 and n Ka2 values obtained for R- and S-propranolol were statistically identical at each temperature (i.e., overlapping within a range of ± 1 S.D.). This result indicated that both types of interactions in the saturable/non-saturable model were essentially non-stereoselective for R/S-propranolol, as is also suggested by data given in Ref. [17]. In addition, the binding parameters measured for the low affinity interactions gave good agreement with values reported in the literature [16,17] when using a model based on a single type of non-saturable interaction.
The nature of the high affinity binding sites was examined further by using comparing the measured binding capacity of this site with the known composition of the HDL support. The total moles of these high affinity sites was consistently in the range of 2.2 to 7.8 nmol for both R- and S-propranolol between 4 and 37°C (average, 4.7 nmol and 5.4 nmol, respectively). It was determined from the protein assay data that approximately 30 (± 2) nmol of immobilized HDL was present in its column. The relatively strong binding of the high affinity sites and the fact that their binding capacity was less than the total moles of HDL strongly suggests that these particular interactions were due to apolipoproteins, for which only a few are present per HDL particle (i.e., up to 5–6 maximum) [7]. The lower binding capacity for these high affinity regions versus the moles of HDL may be due in part to immobilization effects, which can lead to a 50% loss in activity for proteins in the coupling method that was employed in this report [3]. However, it is also possible that only certain types of apolipoproteins may have been involved in this high affinity interaction. The typical levels of apolipoproteins found in HDL are ApoA1 (70%), ApoA2 (20%), and Apo E plus Apo C (10%) [7]. Further research is required in the future to determine if there are indeed only particular apolipoproteins in this group that bind to R/S-propranolol.
The relative importance of the saturable versus non-saturable interactions of R/S-propranolol with HDL was estimated by considering the typical concentrations for each of these agents in clinical samples. The reported therapeutic range for R/S-propranolol in serum is 0.19–0.39 μM [33], and a typical physiological concentration of HDL in serum is 13–14.6 μM [16, 17]. Under these conditions, the high affinity and saturable interactions would be expected from the data in Table 4 to account for 25–40% of the overall binding that occurs between R/S-propranolol and HDL. In the experimental conditions that were employed for the CE studies in Ref. [17], these high affinity sites would have been 85–97% saturated by R/S-propranolol and would have accounted for only 1–10% of the overall measured binding, explaining why these particular interactions were not directly observed in this previous study. A broader range of R/S-propranolol concentrations were used in Ref. [16] in work with equilibrium dialysis, but most of these concentrations were still sufficiently large to have made it difficult to detect the high affinity interactions noted in this current study. In addition, it is important to remember that even in this report a relatively good fit was seen for the data with a single site, non-saturable model (see Figure 5). It was only through a close evaluation and comparison of this data with several interaction models that it became clear that at least two types of interactions were present between R/S-propranolol and HDL.
Frontal analysis studies of verapamil binding to HDL
Verapamil was a second drug used to evaluate binding by the HDL columns. In this case, studies were conducted with racemic verapamil because HDL has been previously reported to have only non-stereoselective interactions with R- and S-verapamil [18]. As indicated earlier, R- and S-propranolol were also found to have non-stereoselective binding to the HDL columns used in this current study.
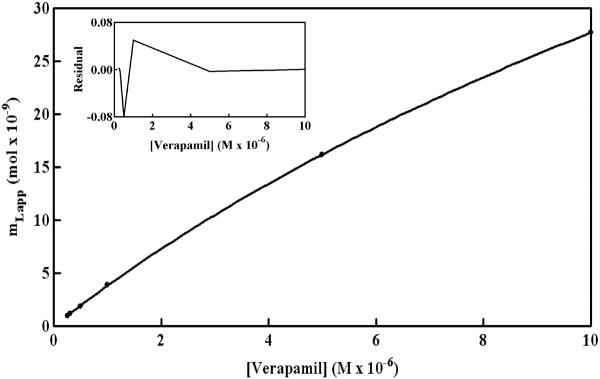
Figure 6 shows a non-reciprocal frontal analysis plot that was obtained for verapamil on an HDL column. These results gave a curved response that deviated from the linear behavior predicted for only non-saturable binding (see equation in Table 2). A correlation coefficient of r = 0.9952 (n = 6) was obtained for the fit of a non-saturable model to these data; however, this model also gave a non-random distribution of data about the best-fit line. A slightly higher correlation coefficient (r = 0.9999) was obtained when using a model based on a single group of saturable sites, but a non-random distribution of data about the best-fit line was also seen in the corresponding residual plot (data not shown).
Figure 6.
Frontal analysis results for the binding of R/S-verapamil to an HDL column at 37°C. These results were obtained in the presence of pH 7.4, 0.067 M potassium phosphate buffer. Other conditions are given in the text. The best-fit line shown is for a model based on a group of non-saturable interactions and a group of saturable sites, which gave a correlation coefficient of 0.99999 (n = 6). The other best-fit parameters are summarized in Table 4.
The model that gave the best-fit for the data in Figure 6 was again based on a group of saturable sites and a set of non-saturable interactions (see summary in Table 4). The correlation coefficient in this case was 0.9999 and only a random distribution of data about the best-fit line was seen in the residual plot. In this fit, the sites responsible for saturable binding had an association equilibrium constant for verapamil of 6.0 (± 2.1) × 104 at 37°C. The non-saturable interaction of verapamil with HDL had an estimated overall affinity of 2.5 (± 1.5) 104 M−1.
These values gave good agreement with the overall affinities of 2.7–2.8 104 M−1 that have been previously reported for the binding of R- and S-verapamil with soluble HDL [18].
A reasonable fit to the data in Figure 6 was also seen for a model in which there were two groups of saturable sites with moderate or lower affinities for verapamil (r = 0.9999). In this model, an association equilibrium constant of 6.3 (± 2.0) × 104 M−1 was obtained for the first group of saturable sites for verapamil on HDL, giving a value similar to that generated by the saturable/non-saturable model. The estimated association equilibrium constant for the second group of saturable sites was 1.5×104 M−1, but this value had a large uncertainty of±2.4 × 106 M−1. This uncertainty plus the non-random distribution of data in the residual plot for the two site saturable model indicated that the saturable/non-saturable model gave a better description of the verapamil/HDL interaction. This conclusion was consistent with the results seen earlier for R- and S-propranolol on the HDL columns.
It is interesting to note that the overall affinities were similar for the non-saturable interactions of verapamil and propranolol with HDL. As discussed in the previous section, this binding is probably the result of interactions between these drugs with phospholipids or the non-polar core of HDL, as has been suggested for both propranolol and verapamil in work with soluble HDL [16–18]. The affinity of verapamil at its saturable sites on HDL was roughly one third the affinity measured for propranolol at its saturable regions, but these two drugs had a much larger difference in the amount of these saturable sites. The amount of saturable sites for verapamil was 54 (± 16) nmol in an HDL column, which was at least 10-times greater than the average of 4.7–5.4 nmol that had been estimated for the saturable sites of R- or S-propranolol. The binding capacity for verapamil at its saturable site was still comparable to the total moles of immobilized HDL in the column (30 nmol) and the expected moles of apolipoproteins that were present [7]. These results indicate that apoliproteins were probably responsible for the saturable binding of verapamil with HDL, as was suggested earlier for propranolol. However, the large difference in the binding capacities for verapamil and propranolol at their saturable sites suggests that different subsets of apolipoproteins or different regions on these apolipoproteins may be interacting with these two drugs.
The typical therapeutic concentration for R/S-verapamil in serum is 0.1–0.2 μM [32]. This information was used along with the binding parameters in Table 4 and the known serum concentration for HDL (13–14.6 μM) to examine the relative importance of saturable versus nonsaturable interactions in the binding of verapamil with HDL. Under normal therapeutic conditions, the saturable interactions would account for about 70–80% of the overall binding between R/S-verapamil and HDL. Under the experimental conditions used in Ref. [18], the saturable sites would have accounted for roughly 40–60% of the overall measured binding. However, the similarity in the affinities for the saturable and non-saturable sites of verapamil, and the fact that only a non-saturable model was considered in Ref. [18], explains why these saturable interactions were not noted in this previous solution-phase study.
CONCLUSIONS
HDL was immobilized in chromatographic columns and examined for use in drug binding studies by HPAC. It was found that this type of column can be employed over four to five days of consistent use without any significant loss of retention for R- and S-propranolol. Frontal analysis experiments with these columns indicated that propranolol and verapamil had two distinct interactions with HDL. One type of interaction had a moderately high affinity and saturable binding and was believed to be due to apolipoproteins. The other interaction was non-saturable and was believed to be due to the phospholipids or non-polar core of HDL. The high affinity sites had association constants of 1.1–1.9 × 105 M−1for R/S-propranolol and 6.0 × 104 M−1 for R/S-verapamil at 37°C. The overall affinity (nKa) for the weaker and non-saturable interactions was 3.7–4.1 × 104 M−1 for R/S-propranolol and 2.5 × 104 M−1 for R/S-verapamil at 37°C, in good agreement with previous solution-phase studies [16,17]. These interactions appeared to be non-stereoselective, giving similar binding parameters for R- and S-propranolol at temperatures ranging from 4 to 37°C.
These results demonstrate the feasibility of using HPAC to study the interactions of HDL with drugs. Some advantages of this approach versus equilibrium dialysis (i.e., the method used in Ref. [16] and a common reference method for drug binding studies) are the ability of HPAC to obtain analysis times of only a few minutes per run and to reuse the same lipoprotein preparation for many experiments. Although the use of CE in drug binding studies requires less ligand for a single analysis, the ability to reuse the ligand in an HPAC column also results in only a small amount of ligand being needed per sample. For instance, the CE studies in Ref. [17] were conducted using approximately 1.5 pmol HDL per injection. In this current report a single HDL column (containing 30 nmol HDL) was used for at least 300 experiments, or an average of 10 pmol HDL per analysis. In addition, the use of the same HDL preparation for multiple studies in HPAC makes it possible with this approach to reduce effects due to batch-to-batch variations in the ligand. The advantage of utilizing HPLC detectors with such columns also made it possible to examine a relatively wide range of drug concentrations. These last two factors made it possible to identify moderately high affinity interactions between HDL and propranolol or verapamil that had not previously been observed when using CE or equilibrium dialysis [16–18]. All of these features indicate that HPAC columns containing immobilized HDL can be useful and powerful tools in the study of drug-lipoprotein interactions.
ACKNOWLEDGEMENTS
This work was supported by the National Institute of Health under grant R01 GM 044931 and was performed in facilities renovated with support under grant RR015468-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Otagiri M. A molecular functional study on the interactions of drugs with plasma proteins. Drug Metab. Pharmacokinet. 2005;20:309–323. doi: 10.2133/dmpk.20.309. [DOI] [PubMed] [Google Scholar]
- 2.Bertucci C, Domenici E. Reversible and covalent binding of drugs to human serum albumin: Methodological approaches and physiological relevance. Curr. Med. Chem. 2002;9:1463. doi: 10.2174/0929867023369673. [DOI] [PubMed] [Google Scholar]
- 3.Hage DS. High-performance affinity chromatography: A powerful tool for studying serum protein binding. J. Chromatogr. B. 2002;768:3–30. doi: 10.1016/s0378-4347(01)00482-0. [DOI] [PubMed] [Google Scholar]
- 4.Hage DS, Jackson A, Sobansky M, Schiel JE, Yoo MJ, Joseph KS. Characterization of drug-protein interactions in blood using high-performance affinity chromatography. J. Sep. Sci. 2009;32:835–853. doi: 10.1002/jssc.200800640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xuan H, Hage DS. Immobilization of α1-acid glycoprotein for chromatographic studies of drug–protein binding. Anal. Biochem. 2005;346:300–310. doi: 10.1016/j.ab.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 6.Mallik R, Xuan H, Guiochon G, Hage DS. Immobilization of α1-acid glycoprotein for chromatographic studies of drug-protein binding. II. Correction for errors in association constant measurements. Anal. Biochem. 2008;376:154–156. doi: 10.1016/j.ab.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonas A. Lipoprotein structure. In: Vance DE, Vance JE, editors. Biochemistry of lipids, lipoproteins and membranes. Elsevier; Amsterdam: 2002. pp. 483–504. [Google Scholar]
- 8.Barklay M. Lipoprotein class distribution in normal and disease states. In: Nelson GJ, editor. Blood lipids and lipoproteins: Quantitation, composition, and metabolism. Wiley; New York: 1972. pp. 587–603. [Google Scholar]
- 9.Wasan KM, Cassidy SM. Role of plasma lipoproteins in modifying the biological activity of hydrophobic drugs. J. Pharm. Sci. 1998;87:411–424. doi: 10.1021/js970407a. [DOI] [PubMed] [Google Scholar]
- 10.Kwong TC. Free drug measurements: Methodology and clinical significance. Clin. Chim. Acta. 1985;151:193–216. doi: 10.1016/0009-8981(85)90082-8. [DOI] [PubMed] [Google Scholar]
- 11.Harmony JAK, Aleson AL, McCarthy BM. Lipoprotein structure. In: Scanu AM, Spector AA, editors. Biochemistry and biology of plasma lipoproteins. Marcel Dekker; New York: 1986. pp. 403–430. [Google Scholar]
- 12.Mbewu AD, Durrington PN. Lipoprotein(a): Structure, properties, and possible involvement in thrombogenesis and atherogenesis. Atherosclerosis. 1990;85:1–14. doi: 10.1016/0021-9150(90)90177-k. [DOI] [PubMed] [Google Scholar]
- 13.Durrington PN. Lipoprotein assembly. In: Durrington PN, editor. Lipoproteins and lipids. Wright; London: 1989. pp. 255–277. [Google Scholar]
- 14.Havel RJ, Kane JP. Structure and metabolism of plasma lipoproteins. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The metabolic and molecular basis of inherited disease. McGraw-Hill; New York: 1995. pp. 1129–1138. [Google Scholar]
- 15.Skipski VR. Lipid composition of lipoproteins in normal and diseased states. In: Nelson GJ, editor. Blood lipids and lipoproteins: Quantitation, composition, and metabolism. Wiley; New York: 1972. pp. 471–583. [Google Scholar]
- 16.Glasson S. The distribution of bound propranolol between the different human serum proteins. Mol. Pharmacol. 1980;17:187–191. [PubMed] [Google Scholar]
- 17.Ohnishi T, Mohamed NAL, Shibukawa A, Kuroda Y, Nakagawa T, El Gizawy S, Askal HF, El Kommos ME. Frontal analysis of drug-plasma lipoprotein binding using capillary electrophoresis. J. Pharm. Biomed. Anal. 2002;27:607–614. doi: 10.1016/s0731-7085(01)00569-6. [DOI] [PubMed] [Google Scholar]
- 18.Mohamed NAL, Kuroda Y, Shibukawa A, Nakagawa T, El Gizawy S, Askal HF, El Kommos ME. Enantioselective binding analysis of verapamil to plasma lipoproteins by capillary electrophoresis-frontal analysis. J. Chromatogr. A. 2000;875:447–453. doi: 10.1016/s0021-9673(99)01288-1. [DOI] [PubMed] [Google Scholar]
- 19.Mohamed NAL, Kuroda Y, Shibukawa A, Nakagawa T, El Gizawy S, Askal HF, El Kommos ME. Binding analysis of nilvadipine to plasma lipoproteins by capillary electrophoresis-frontal analysis. J. Pharm. Biomed. Anal. 1999;21:1037–1043. doi: 10.1016/s0731-7085(99)00197-1. [DOI] [PubMed] [Google Scholar]
- 20.Kim HS, Wainer IW. Rapid analysis of the interactions between drugs and human serum albumin (HSA) using high-performance affinity chromatography (HPAC) J. Chromatogr. B. 2008;870:22–26. doi: 10.1016/j.jchromb.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollósy F, Valkó K, Hersey A, Nunhuck S, Kéri G, Bevan C. Estimation of volume of distribution in humans from high throughput HPLC-based measurements of human serum albumin binding and immobilized artificial membrane partitioning. J. Med. Chem. 2006;49:6958–6971. doi: 10.1021/jm050957i. [DOI] [PubMed] [Google Scholar]
- 22.Buchholz L, Cai C-H, Andress L, Cleton A, Brodfuehrer J, Cohen L. Evaluation of the human serum albumin column as a discovery screening tool for plasma protein binding. Eur. J. Pharm. Sci. 2002;15:209–215. doi: 10.1016/s0928-0987(01)00219-6. [DOI] [PubMed] [Google Scholar]
- 23.Loun B, Hage DS. Chiral separation mechanisms in protein-based HPLC columns. 1. Thermodynamic studies of (R)- and (S)-warfarin binding to immobilized human serum albumin. Anal. Chem. 1994;66:3814–3822. doi: 10.1021/ac00093a043. [DOI] [PubMed] [Google Scholar]
- 24.Hage DS, Chen J. Quantitative affinity chromatography: Practical aspects. In: Hage DS, editor. Handbook of affinity chromatography. CRC Press/Taylor & Francis; New York: 2006. pp. 595–628. [Google Scholar]
- 25.Tweed S, Loun B, Hage DS. Effects of ligand heterogeneity in the characterization of affinity columns by frontal analysis. Anal. Chem. 1997;69:4790–4798. doi: 10.1021/ac970565m. [DOI] [PubMed] [Google Scholar]
- 26.Mallik R, Xuan H, Hage DS. Development of an affinity silica monolith containing α1-acid glycoprotein for chiral separations. J. Chromatogr. A. 2007;1149:294–304. doi: 10.1016/j.chroma.2007.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 28.Wiechelman KJ, Braun RD, Fitzpatrick JD. Investigation of the bicinchoninic acid protein assay: Identification of the groups responsible for color formation. Anal. Biochem. 1988;175:231–237. doi: 10.1016/0003-2697(88)90383-1. [DOI] [PubMed] [Google Scholar]
- 29.Richmond W. Preparation and properties of a cholesterol oxidase from Nocardia sp. and its application to the enzymatic assay of total cholesterol in serum. Clin. Chem. 1973;19:1350–1356. [PubMed] [Google Scholar]
- 30.Allain CC, Poon LS, Chan CSG, Richmond W, Fu PC. Enzymatic determination of total serum cholesterol. Clin. Chem. 1974;20:470–475. [PubMed] [Google Scholar]
- 31.Kim HS, Mallik R, Hage DS. Chromatographic analysis of carbamazepine binding to human serum albumin. II. Comparison of the Schiff base and N-hydroxysuccinimide immobilization methods. J. Chromatogr. B. 2006;837:138–146. doi: 10.1016/j.jchromb.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 32.Mallik R, Yoo MJ, Chen S, Hage DS. Studies of verapamil binding to human serum albumin by high-performance affinity chromatography. J. Chromatogr. B. 2008;876:69–75. doi: 10.1016/j.jchromb.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tietz NW, editor. Textbook of Clinical Chemistry. Saunders; Philadelphia: 1986. [Google Scholar]