Abstract
Current therapies for glioblastoma (GBM) target bulk tumor through measures such as resection and radiotherapy. However, recent evidence suggests that targeting a subset of tumor cells, so-called cancer stem cells, may be critical for inhibiting tumor growth and relapse. The subventricular zone (SVZ), which lines the ventricles of the brain, is thought to be the origin for the majority of neural stem cells and potentially cancer stem cells. Therefore, we assessed the relationship between tumor contact with the SVZ as determined by MRI, cancer stem cell gene expression and survival in 47 patients with GBM. Using DNA microarrays, we found that genes associated with cancer stem cells were not over-expressed in tumors contacting the SVZ. Contact with the SVZ trended with shorter survival (median 358 versus 644, P = 0.066). Over-expression of CD133 (prominin-1) and maternal embryonic leucine zipper kinase (MELK) was associated with shorter survival, whereas mitogen activated protein kinase 8 (MAPK8) was associated with longer survival (P values 0.008, 0.005 and 0.002 respectively). Thus we found no evidence of a stem-cell derived genetic signature specific for GBM in contact with the SVZ, but there was a relationship between stem cell gene expression and survival. More research is required to clarify the relationship between the SVZ, cancer stem cells and survival.
Keywords: Glioblastoma, Glioma, Subventricular zone, Microarray, Stem cell, Survival, Genomics, MRI
Introduction
Survival of patients with glioblastoma (GBM) remains poor. Current therapy for gliomas is typically based on cytoreductive strategies, such as surgery and radiotherapy, which target the bulk of the tumor mass rather than specific cell types [1]. Recently it has been suggested that a minority of the cells in the tumor, so-called cancer stem cells, account for most of the proliferation, growth and radioresistance [2; reviewed in 3]. These cells can also propagate tumors in vitro. Therefore the elimination of these cancer stem cells may prove necessary to effect a cure for glioma patients [4]. The subventricular zone (SVZ), which lines the ventricles, maintains the ability to generate neurons and glia throughout adulthood, and in animal models, gliomas can be generated from this cell population. It has been suggested that the SVZ is a source for cancer stem cells that initiate gliomagenesis [5].
GBM are heterogeneous tumors, histopathologically, radiographically and genetically [6]. Several genetic subtypes have been characterized that are associated with varying survival rates. For instance, an oligodendroglioma-like genetic signature is associated with longer survival [7, 8]. Survival also may be impacted by the tumor’s relationship to the SVZ [9, 10]. Lim et al. [9] have suggested that GBM contacting the SVZ are more likely to recur as multifocal disease. And previously it has been shown that multifocal disease is associated with shortened survival [11]. It also has been proposed that GBM contacting the SVZ may be more likely to be stem-cell-derived, with potential impact on treatment decisions [9].
Recently several stem cell marker proteins have been identified. The most well-characterized, CD133, is associated with shortened survival [12]. However, to date, there has been no analysis of gene expression and survival in tumors based on contact with the SVZ. Therefore, we used microarrays to analyze the relationship between tumors associated with the SVZ, expression of cancer stem cell and proliferative genes, and survival.
Materials and methods
Patient selection
All patients participating in this study signed institutional review board-approved informed consent. Data acquisition was performed in compliance with all applicable Health Insurance Portability and Accountability Act (HIPAA) regulations. Patients were retrospectively selected from our institution’s neuro-oncology database. All patients (n = 47) who met the following criteria were selected: (1) pathology confirmed GBM (2) baseline (pre-surgical) MRI (3) fresh-frozen tissue adequate for microarray analysis. All patients were treated with radiation therapy (RT; 6000 ± 500 cGy) and subtotal/total resection at time of initial tumor presentation. Accrual dates for this study were October 1999 to January 2004. A total of 26/47 (55.3%) patients received adjuvant chemotherapy, most commonly temozolomide (15/47, 31.9%). Some patients also received salvage chemotherapy.
Imaging
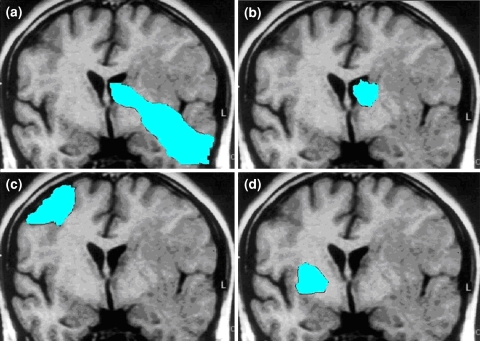
MRI sequences were acquired on a 1.5T scanner and typically included axial T1 weighted (TR 400, TE 15, slice thickness 5 mm), T2 weighted fast spin-echo (TR 4000, TE 126-130, slice thickness 5 mm), diffusion-weighted and gadopentetate dimeglumine enhanced (Magnevist; Berlex, Wayne, NJ; 0.1 mmol/kg) axial and coronal T1 weighted images (TR 400, TE 15, slice thickness 3 mm), with a field of view of 24 cm and a matrix size of 256 × 256. Post-contrast images were acquired immediately after contrast injection. The contrast-enhancing tumor was assessed for contact with the SVZ and cortex as described in [9]. Tumors were segregated into 4 groups by a neuroradiologist (W.P.): group I, tumor contacting SVZ and infiltrating cortex 9 (n = 13); group II, tumor contacting SVZ but not involving the cortex (n = 12); group III, tumor involving cortex but not contacting the SVZ (n = 16); group IV, tumor involving neither SVZ nor cortex (n = 6; Fig. 1). A minority of patients had multifocal disease at initial presentation (7/47, 15%). For these patients, the scans were graded according to the dominant tumor nodule.
Fig. 1.
Tumor grading. Group I (a), tumor contacting SVZ and infiltrating cortex (n = 13); group II (b), tumor contacting SVZ but not involving the cortex (n = 12); group III (c), tumor involving cortex but not contacting the SVZ (n = 16); group IV (d), tumor involving neither SVZ nor cortex (n = 6)
Microarray data
RNA was purified from fresh frozen tumor biopsies and was hybridized to U133A and U133 Plus 2.0 Arrays at our DNA Microarray Facility. The data files generated were normalized using RMA from Bioconductor (for R, version 2.6; http://cran.r-project.org/bin/windows/base/), relative to other Affymetrix microarrays of the same platform using the Celsius Database (http://www.ncbi.nlm.nih.gov/pubmed/17570842). Only probes from the U133A portion that were retained in U133 Plus 2.0 arrays were analyzed for each sample. RMA normalized data was imported into dChip [12] for differential expression analysis among genes in different SVZ groups. We compared the fold change for the mean of each gene in the different SVZ groups, and used a t-test to assess the significance. Significant genes were further analyzed by the DAVID bioinformatics tool (The US National Institutes of Health (NIH) Database for Annotation, Visualization and Integrated Discovery; http://david.abcc.ncifcrf.gov; [13, 14]), to identify enriched biological groups [15].
Heat map generation
Gene expression was visualized through heat maps generated by dChip. Differentially expressed genes identified among the SVZ groups were hierarchically clustered, while the samples were arranged by SVZ group.
Statistical methods
The survival library from R (version 2.6; http://cran.r-project.org) and the Cox proportional hazards model were used to analyze the relationship between gene expression and survival. Results for all genes were obtained and the values for the stem cell genes of interest, and the differentially expressed genes for the SVZ groups were extracted. Cox model regression analysis results were obtained for each of the genes and a transformed hazard ratio (Z score, the ratio of the regression coefficient to the standard error) was generated. A positive score Z coefficient indicates higher expression corresponding to higher hazard rate and shorter survival, and lower expression corresponding to a lower hazard rate and longer survival. Kaplan–Meier curves with log-rank test were used to analyze survivability patterns between SVZ grades and gene expression. Expression fold change between tumor groups was computed with a fold change threshold of 1.2 with a lower 90% confidence bound, and a t-test P value of 0.05. For most analyses, a P value of <0.05 was accepted as significant. Some analyses were performed with a Bonferroni correction for multiple testing.
Results
Poor correlation of stem cell gene expression
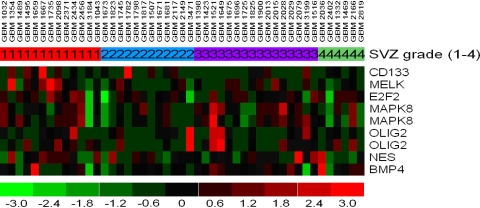
A total of 7 genes of interest related to cancer stem cells or tumor proliferation, based on previous literature [12, 16–20], were selected for analysis. The genes were CD133 (prominin1), maternal embryonic leucine zipper kinase (MELK), bone morphogenetic protein 4 (BMP 4), E2F transcription factor 2 (E2F 2), mitogen-activated protein kinase 8 (MAPK 8; also known as Jun NH2-terminal kinase), OLIG2, and nestin (NES). We attempted to find a stem cell signature which consisted of a number of genes whose expression correlated highly with one another or over expression of stem cells genes in a specific tumor group, but found none. The correlation matrix showed very low correlation between any of the stem cell genes of interest (Table 1). Lack of correlation is confirmed on the heat map (Fig. 2).
Table 1.
Correlation of stem cell gene expression in 47 patients with GBM
| CD133 | MELK | E2F2 | MAPK8 (P 1a) | MAPK8 (P 2) | BMP4 | OLIG2 (P 1) | OLIG2 (P 2) | NES | |
|---|---|---|---|---|---|---|---|---|---|
| CD133 | – | 0.22 | −0.17 | −0.21 | −0.11 | −0.24 | −0.09 | −0.06 | 0.04 |
| MELK | 0.22 | – | 0.22 | −0.03 | 0.06 | 0.04 | −0.14 | −0.07 | 0.36 |
| E2F2 | −0.17 | 0.22 | – | 0.53 | 0.52 | 0.53 | −0.16 | −0.20 | 0.06 |
| MAPK8 (PS 1) | −0.21 | −0.03 | 0.53 | – | 0.81 | 0.43 | 0.13 | 0.22 | −0.02 |
| MAPK8 (PS 2) | −0.11 | 0.06 | 0.52 | 0.81 | – | 0.27 | 0.17 | 0.13 | −0.01 |
| BMP4 | −0.24 | 0.04 | 0.53 | 0.43 | 0.27 | – | −0.17 | −0.13 | 0.15 |
| OLIG2 (PS 1) | −0.09 | −0.14 | −0.16 | 0.13 | 0.17 | −0.17 | – | 0.77 | −0.05 |
| OLIG2 (PS 2) | −0.06 | −0.07 | −0.20 | 0.22 | 0.13 | −0.13 | 0.77 | – | −0.11 |
| NES | 0.04 | 0.36 | 0.06 | −0.02 | −0.01 | 0.15 | −0.05 | −0.11 | – |
a P probe. Both MAPK8 and OLIG2 had 2 probes available for analysis
Fig. 2.
Heat map of stem cell probes across group I–IV. Red corresponds to higher gene expression levels. Patient samples noted along top row
Stem cell genes associated with survival
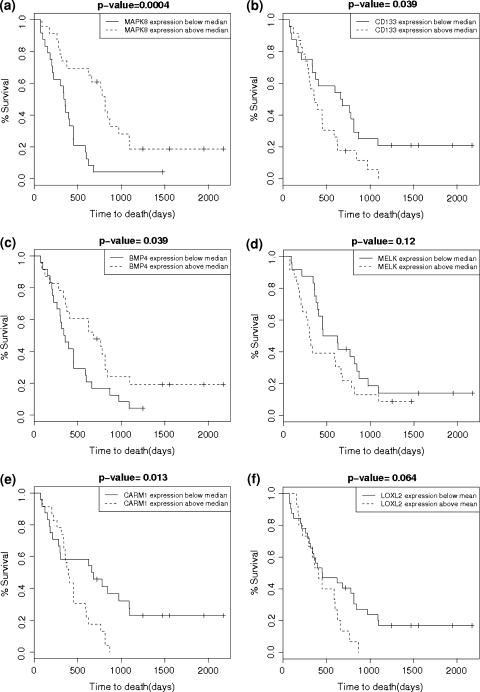
In the survival analysis (Cox Model), 6 of the 7 genes of interest had at least one probe with a P value significant at the 0.05 level. With a multiple testing correction for the 9 probes, 2 of these genes (MAPK8 and MELK) are significantly associated with survival at a Bonferroni adjusted P value of 0.006 (Table 2). Increased expression levels of CD133 and MELK were associated with shorter survival, whereas increased expression levels of nestin, OLIG2, BMP4, ETF2 and MAPK8 were associated with longer survival.
Table 2.
Stem cell/proliferative gene expression and survival
| Gene | Za | P valueb,c | |
|---|---|---|---|
| Gene symbol | Probe | ||
| MAPK8 | 210477_x_at | −3.17 | 0.002 |
| MAPK8 | 210671_x_at | −2.46 | 0.014 |
| BMP4 | 211518_s_at | −2.11 | 0.035 |
| NES | 218678_at | −2.11 | 0.035 |
| OLIG2 | 213825_at | −1.98 | 0.048 |
| OLIG2 | 213824_at | −1.51 | 0.130 |
| E2F2 | 207042_at | −1.36 | 0.170 |
| CD133 | 204304_s_at | 2.65 | 0.008 |
| MELK | 204825_at | 2.79 | 0.005 |
a Z is the transformed hazard ratio. A negative Z score indicates higher expression levels are correlated with longer survival, and a positive Z score indicates higher expression is correlated with shorter survival
b P value from Cox model
cValues <0.05 bolded
Differentially expressed genes in group II tumors: association with immune system not cancer stem cells
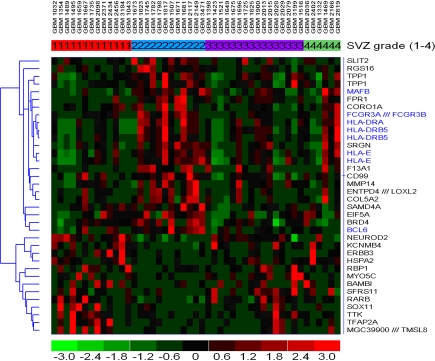
Although we did not find that stem cell gene expression was increased in tumors contacting the SVZ, several other genes were over-expressed in tumors contacting the SVZ. The list of differentially expressed genes was submitted to the DAVID bioinformatics tool in order to look at over represented functional groups and to determine general function. Of the 33 genes (36 probes) over-expressed in group II tumors, 7 showed a Gene Ontology (GO) term of the immune system process (P value = 0.03). These non stem-cell genes were: FCGR3A, HLA-DRB5, BCL6, FCGR3B, MAFB, HLA-DRA, HLA-E. These genes all had higher expression in the group II, with a fold change ranging from 1.48 to 2.43 (Table 3). The average fold change of the 7 immune system differentially expressed genes was 1.82. Figure 3 is a heat map showing gene expression by SVZ grade. Note the high expression of these immune-related genes in group II tumors. We also assessed the expression of these genes in group IV versus groups I and III, since group IV tumors also lack cortical involvement. Except for BCL6, the immune genes were all over-expressed in group IV tumors with fold changes ranging from 1.31 to 2.31.
Table 3.
Immune system genes over-expressed in group II tumors
| Gene symbol | Fold change | 90% CI | P valuea,b |
|---|---|---|---|
| FCGR3A | 2.43 | 1.49–3.73 | 0.020 |
| BCL6 | 2.05 | 1.43–2.80 | 0.013 |
| HLA-DRA | 1.79 | 1.20–2.51 | 0.039 |
| HLA-DRB5 (PS 2) | 1.78 | 1.20–2.54 | 0.037 |
| HLA-DRB5 (PS 1) | 1.77 | 1.21–2.48 | 0.033 |
| MAFB | 1.67 | 1.25–2.23 | 0.011 |
| HLA-E (PS 1) | 1.56 | 1.27–1.93 | 0.002 |
| HLA-E (PS 2) | 1.48 | 1.24–1.76 | 0.002 |
PS probes
a P value from Cox model
bValues <0.05 bolded
Fig. 3.
Heat map of differentially expressed genes between group II tumors versus the remainders. Note that several immune related genes (blue) show higher expression in group 2 tumors. Red corresponds to higher gene expression levels
Association of genes overexpressed in SVZ-contacting tumors and survival
We also analyzed the relationship between genes over-expressed in group II tumors and survival (Table 4). Expression of 5 genes (either increased or decreased) correlated with shorter survival. Comparing gene expression in tumors grade I and II versus III and IV (i.e., all tumors that contact the SVZ versus all that do not), led to the identification of CARM1 which is highly expressed in SVZ-contacting tumors and also is highly correlated with survival (P = 0.005). Thus this analytic approach may be of use in screening for genes that may impact the relationship between SVZ contact and survival.
Table 4.
Genes over-expressed in group II tumors that correlate with survival
| Gene symbol | Za | P value | Fold change | 90% CIb | P value FCc,d,e |
|---|---|---|---|---|---|
| COL5A2 | 3.15 | 0.002 | 2.03 | 1.30–3.11 | 0.026 |
| LOXL2 | 3.10 | 0.002 | 2.29 | 1.31–3.57 | 0.046 |
| SLIT2 | 2.42 | 0.016 | 2.04 | 1.30–2.99 | 0.034 |
| SOX11 | 2.37 | 0.018 | −2.74 | −1.37–8.14 | 0.024 |
| TTK | 2.32 | 0.020 | −1.61 | −1.23–2.07 | 0.010 |
a Z is the transformed hazard ratio. A negative Z score indicates higher expression levels are correlated with longer survival, and a positive Z score indicates higher expression is correlated with shorter survival
b CI Confidence interval
c FC fold change
d P value from Cox model
eValues <0.05 bolded
Kaplan–Meier analysis
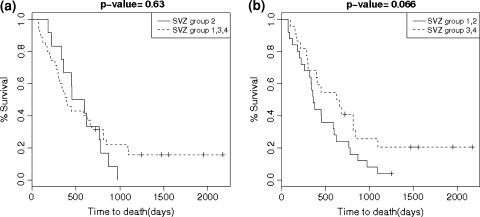
We generated Kaplan–Meier curves to compare survival between tumors in group I and II versus III and IV. There was a trend towards shorter survival (median survival 358 versus 654 days, P = 0.066; Fig. 4). There was no significant difference in survival curves for group II versus I, III, IV (Fig. 4). There was no statistically significant difference between rates of multifocal disease, at presentation and recurrence, between groups. However, there was a significant difference between gross total resection rates in group I versus remainder tumors (Table 5). Figure 5 shows Kaplan–Meier curves for patients grouped by gene expression.
Fig. 4.
Kaplan–Meier curves of group II versus groups I, III and IV combined (a) and Kaplan–Meier curves of group I and II combined versus groups III and IV combined (b)
Table 5.
Clinical data by tumor group
| Group | I | II | III | IV |
|---|---|---|---|---|
| n | 13 | 12 | 16 | 6 |
| Age (SEa) | 54 (4.5) | 48.8 (2.5) | 50.6 (3.7) | 44 (4.5) |
| KPSb (SE) | 83.8 (4.6) | 80.8 (5.1) | 85 (4.2) | 88 (7.3) |
| Adj chemoc (%) | 6/13 (46.2) | 8/12 (66.7) | 9/16 (56.3) | 3/6 (50.0) |
| Temozolomided (%) | 2/13 (15.4) | 4/12 (33.3) | 7/16 (43.8) | 2/6 (33.3) |
| GTRe,f (%) | 2/13 (15.4) | 6/12 (50.0) | 7/16 (43.8) | 4/6 (66.7) |
| Multifocal, initial (%) | 1/13 (7.7) | 1/12 (8.3) | 4/16 (25.0) | 1/6 (16.7) |
| Multifocal, relapseg (%) | 3/11 (27.3) | 1/8 (12.5) | 4/13 (30.8) | 1/5 (20.0) |
a SE standard error
b KPS Karnosfsky performance status at presentation
c adj chemo adjuvant chemotherapy of any kind
d temozolomide adjuvant chemotherapy with temozolomide
e GTR gross total resection
f P = 0.03 for group I versus other tumors, chi-square test
gSix patients have yet to progress, and follow-up scans were not available for 4 patients, accounting for reduced patient number for this variable
Fig. 5.
Kaplan–Meier curves for selected genes: a MAPK8, b CD133, c BMP-4, d MELK, e CARM1 and f LOXL2. For all graphs the dashed line indicates survival data for patients with greater than median gene expression levels, and the solid line is for patients with less than median gene expression levels. Note that increased levels of MAPK and BMP-4 expression are associated with longer survival, whereas increased CD133 and MELK expression levels correlate with shorter survival. Increased CARM1 and LOXL2 (enriched in SVZ-contacting tumors) also are correlated with poorer survival
Discussion
It has been hypothesized that gliomas arise from stem cells originating in the SVZ. Previously it has been reported that more aggressive patterns of GBM recurrence are associated with tumors that contact the SVZ, and that these tumors may express a “stem-cell-derived” phenotype that could impact treatment decisions [9]. Therefore we analyzed the relationship between expression of cancer stem-cell genes, contact of tumor with the SVZ and patient survival.
Similar to previous work [9, 10], we found that patients with GBM which contacted the SVZ trended with shorter survival. One possibility is that shortened survival may be related to tumor size, as tumors that contact both the cortex and SVZ tend to be large. However, previous reports have shown little correlation between GBM size and survival [11]. Additionally, Barami et al. [21] found that contact with the ventricular wall was independent of size.
Another possibility is that tumors that contact the SVZ have higher rates of multifocal or subependymal/leptomeningeal spread. We did not find a significant difference in rates of non-local recurrence between tumor groups based on SVZ contact. And only one patient had leptomeningeal recurrence. Unlike Lim et al. [9] we found that group III tumors had the highest rates of multifocal disease, both at initial presentation and at recurrence. Thus it is unclear if tumors that contact the SVZ have more aggressive patterns of recurrence. Small sample sizes may account for these discordant results.
There was a difference in gross total resection rates between groups. Group I tumors (those that contact both the ventricle and cortex) had a lower gross total resection rate than other tumors. Since complete resection is associated with better survival [22], this could be a potential confound for the survival data. Additionally, deeper tumors may have more impact on critical structures, increasing their impact on disability. Thus we cannot say whether tumors that contact the SVZ are intrinsically more malignant than those that do not.
We found no relationship between stem-cell gene expression and SVZ grade. Specifically there was no correlation in expression between the selected stem cell genes, and there was no enrichment of stem cell genes in tumors that contacted the SVZ. Although not correlated with tumor location, expression of several stem cell genes, including CD133, was associated with shortened survival, similar to previous reports [12, 23]. For instance, Zhang et al. [23] found that CD133 expression was negatively correlated with both overall survival and progression-free survival. CD133 expression has been shown to be associated with resistance to radiation and chemotherapy [24]. Similar results have been demonstrated in mice models of GBM [25]. Interestingly, BMP4 has been hypothesized to down-regulate CD133 and increase survival in a mouse model [26]. Although there was no significant correlation between CD133 and BMP4 expression, increased BMP4 expression was associated with longer survival.
Unlike the report of Zhang et al. [23] we found that higher nestin expression was associated with longer survival. Some differences between the two experiments are notable. Zhang measured protein level with immunohistochemistry, whereas we examined gene expression. Zhang also looked at grade II–IV tumors, whereas we restricted our analysis to grade IV tumors (GBM). Another group [27] found no difference in survival based on nestin expression levels. Similarly, increased OLIG2 expression was associated with longer survival. OLIG2 is associated with oligodendrogliomal lineage development [17], and an oligodendroglioma component and genetic signature have previously been shown to be associated with longer survival [28]. Another gene whose expression was associated with longer survival was MAPK8 (also known as JNK). Previously it has been shown that in cultured glioma cells, inhibition of this kinase reduces VEGF secretion [29]. Since VEGF is associated with malignancy, this may explain why patients with tumors with lower MAPK8 gene expression had longer survival.
Since patients in group I + II trended with shorter survival, we also analyzed the expression of genes in SVZ-contacting tumors compared to those that did not contact the SVZ in order to look for candidate genes that may be involved with shorter survival. Several genes, including CARM1 and LOXL2, over-expressed in SVZ-contacting tumors, were associated with poor survival. LOXL2 has been shown to promote cell migration in breast cancer [30], and is associated with a poor prognosis in squamous cell carcinomas [31], but this is the first report of an association with poor survival in GBM. CARM1 promotes breast cancer proliferation through an estrogen-dependent mechanism [32], but has no known role in glioma malignancy.
Interestingly, although no stem-cell gene expression signature was associated with SVZ group II, many genes associated with the immune system were over-expressed in group II, including several MHC genes. This is of potential importance as MHC expression modulates response to dendrictic cell therapy currently being developed to treat GBM, and thus tumors that contact the SVZ may respond differently to this treatment compared to other tumors [33]. Additionally, it has been shown that migrating glioma cells down-regulate MHC class II genes. If the SVZ is the origin for gliomagenic cells, this suggests that high expression of these genes may account for the lack of spread of group II tumor progenitor cells away from the SVZ prior to tumor formation [34].
In conclusion, we found no evidence that tumors contacting the SVZ were more likely, based on gene expression, to be “stem-cell-derived”, than tumors not contacting the SVZ. Two possible explanations are: (1) the SVZ is not the origin for tumor producing cells; or (2) cells responsible for gliomagenesis are produced in the SVZ but migrate towards the cortex before tumorigenesis. The second hypothesis is supported by animal studies which have shown that gliomas arising from the periventricular region can grow along white matter tracts, losing their connection to the SVZ [35]. We did find, however, that SVZ-contacting cells over-expressed several genes associated with patient survival and with the immune system. The expression of immune-related genes could impact response to treatment, and may mediate cell migration away from the SVZ. Thus, more research is required to clarify the relationship between migration of SVZ derived cancer stem cells and the temporal relationship to gliomagenesis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Melanie Kappadakunnel and Ascia Eskin contributed equally to this report.
References
- 1.Newton HB. Glioblastoma multiforme. Curr Treat Options Neurol. 2008;10:285–294. doi: 10.1007/s11940-008-0031-z. [DOI] [PubMed] [Google Scholar]
- 2.Bao S, Wu Q, Mc Lendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 3.Stiles CD, Rowitch CH. Glioma stem cells: a midterm exam. Neuron. 2008;58:834–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 4.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumourigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 5.Gil-Perotin S, Marin-Husstege M, Li J, et al. Loss of p53 induces changes in the behavior of subventricular zone cells: implication for the genesis of glial tumors. J Neurosci. 2006;26:1107–1116. doi: 10.1523/JNEUROSCI.3970-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rees JH, Smirniotopoulos JG, Jones RV. Glioblastoma multiforme: radiologic-pathologic correlation. Radiographics. 1996;16:1413–1438. doi: 10.1148/radiographics.16.6.8946545. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Scheck AC, Cloughesy TF, et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pope WB, Chen JH, Dong J, et al. Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology. 2008;249:268–277. doi: 10.1148/radiol.2491072000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim DA, Cha S, Mayo MC, Chen MH, Keles E, Vandenberg S, Berger MS. Relationship of glioblastoma multiforme to neural stem cell regions predicts invasive and multifocal tumor phenotype. Neuro Oncol. 2007;9:424–428. doi: 10.1215/15228517-2007-023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaichana KL, McGirt MJ, Frazier J, et al. Relationship of glioblastoma multiforme to the lateral ventricles predicts survival following tumor resection. J Neurooncol. 2008;89:219–224. doi: 10.1007/s11060-008-9609-2. [DOI] [PubMed] [Google Scholar]
- 11.Pope WB, Sayre J, Perlina A, Villablanca JP, Mischel PS, Cloughesy TF. MR imaging correlates of survival in patients with high-grade gliomas. AJNR Am J Neuroradiol. 2005;10:2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 12.Pallini R, Ricci-Vitiani L, Banna GL, et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14:8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- 13.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 14.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4(5):P3. doi:10.1186/gb-2003-4-5-p3 [PubMed]
- 15.Theodorou V, Kimm MA, Boer M, Wessels L, Theelen W, Jonkers J, Hilkens J. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet. 2007;39:759–769. doi: 10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- 16.Mangiola A, Lama G, Giannitelli C, et al. Stem cell marker nestin and c-Jun NH2-terminal kinases in tumor and peritumor areas of glioblastoma multiforme: possible prognostic implications. Clin Cancer Res. 2007;13:6970–6977. doi: 10.1158/1078-0432.CCR-07-1229. [DOI] [PubMed] [Google Scholar]
- 17.Ligon KL, Huillard E, Mehta S, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto OK, Oba-Shinjo SM, Lopes L, Nagahashi-Marie SK. Expression of HOXC9 and E2F2 are up-regulated in CD133+ cells isolated from human astrocytomas and associate with transformation of human astrocytes. Biochim Biophys Acta. 2007;1769:437–442. doi: 10.1016/j.bbaexp.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Nakano I, Masterman-Smith M, Saigusa K, et al. Maternal embryonic leucine zipper kinase is a key regulator of the proliferation of malignant brain tumors, including brain tumor stem cells. J Neurosci Res. 2008;86:48–60. doi: 10.1002/jnr.21471. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Son MJ, Woolard K, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barami K, Sloan AE, Rojiani A, Schell MJ, Staller A, Brem A. Relationship of gliomas to the ventricular walls. J Clin Neurosci. 2009;16:195–201. doi: 10.1016/j.jocn.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 22.McGirt MJ, Chaichana LK, Gathinji M, et al. Independent association of extent of resection with survival in patients with malignant brain astrocytoma. J Neurosurg. 2009;110:156–162. doi: 10.3171/2008.4.17536. [DOI] [PubMed] [Google Scholar]
- 23.Zhang M, Song T, Yang L, Chen R, Wu L, Fang J. Nestin and CD 133: valuable stem cell-specific markers for determining clinical outcome of glioma patients. J Exp Clin Caner Res. 2008;27:85. doi: 10.1186/1756-9966-27-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related “self renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3023. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 25.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 26.Piccirillo SG, Reynolds BA, Zanetti N, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 27.Chinnaiyan P, Wang M, Rojiani AM, et al. The prognostic value of nestin expression in newly diagnosed glioblastoma: report from the Radiation Therapy Oncology Group. Radiat Oncol. 2008;3:32. doi: 10.1186/1748-717X-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salvati M, Formichella AI, D’Elia A (2009) Cerebral glioblastoma with oligodendrogliomal component: analysis of 36 cases. J Neurooncol, Apr 3 [Epub ahead of print] [DOI] [PubMed]
- 29.Yoshino Y, Aoyagi M, Tamaki M, Duan L, Morimoto T, Ohno K. Activation of p38 MAPK and/or JNK contributes to increased levels of VEGF secretion in human malignant glioma cells. Int J Oncol. 2006;29:981–987. [PubMed] [Google Scholar]
- 30.Hollosi P, Yakushiji JK, Fong KS, Csiszar K, Fong SF (2009) Lysyl oxidase-like 2 promotes migration in noninvasive breast cancer cells but not in normal breast epithelial cells. Int J Cancer, Feb 3 [Epub ahead of print] [DOI] [PubMed]
- 31.Peinado H, Moreno-Bueno G, Hardisson D, et al. Lysyl oxidase-like 2 as a new poor prognosis marker of squamous cell carcinomas. Cancer Res. 2008;68:4541–4550. doi: 10.1158/0008-5472.CAN-07-6345. [DOI] [PubMed] [Google Scholar]
- 32.Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–306. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- 33.Liau LM, Prins RM, Kiertscher SM, et al. Dendritic cell vaccination in glioblastoma patients induces systemic and intracranial T-cell responses modulated by the local central nervous system tumor microenvironment. Clin Cancer Res. 2005;11:5515–5525. doi: 10.1158/1078-0432.CCR-05-0464. [DOI] [PubMed] [Google Scholar]
- 34.Zagzag D, Salnikow K, Chiriboga L, et al. Downregulation of major histocompatibility complex antigens in invading glioma cells: stealth invasion of the brain. Lab Invest. 2005;85:328–341. doi: 10.1038/labinvest.3700233. [DOI] [PubMed] [Google Scholar]
- 35.Zhu Y, Guignard F, Zhao D, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF 1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]