Abstract
Skeletal muscle adaptations to exercise confer many of the health benefits of physical activity and occur partly through alterations in skeletal muscle gene expression. The exact mechanisms mediating altered skeletal muscle gene expression in response to exercise are unknown. However, in recent years, chromatin remodelling through epigenetic histone modifications has emerged as a key regulatory mechanism controlling gene expression in general. The purpose of this study was to examine the effect of exercise on global histone modifications that mediate chromatin remodelling and transcriptional activation in human skeletal muscle in response to exercise. In addition, we sought to examine the signalling mechanisms regulating these processes. Following 60 min of cycling, global histone 3 acetylation at lysine 9 and 14, a modification associated with transcriptional initiation, was unchanged from basal levels, but was increased at lysine 36, a site associated with transcriptional elongation. We examined the regulation of the class IIa histone deacetylases (HDACs), which are enzymes that suppress histone acetylation and have been implicated in the adaptations to exercise. While we found no evidence of proteasomal degradation of the class IIa HDACs, we found that HDAC4 and 5 were exported from the nucleus during exercise, thereby removing their transcriptional repressive function. We also observed activation of the AMP-activated protein kinase (AMPK) and the calcium–calmodulin-dependent protein kinase II (CaMKII) in response to exercise, which are two kinases that induce phosphorylation-dependent class IIa HDAC nuclear export. These data delineate a signalling pathway that might mediate skeletal muscle adaptations in response to exercise.
Introduction
Skeletal muscle is a highly plastic tissue and its phenotype plays a key role in human health and performance. Aerobic exercise induces skeletal muscle adaptations that confer many of the benefits of physical activity. Alterations in gene expression in response to exercise contribute to these adaptations (Mahoney et al. 2005). The exact molecular mechanisms that mediate exercise-induced gene expression have not been fully elucidated. In recent years, the importance of chromatin remodelling in regulated gene expression has become evident (Strahl & Allis, 2000). Chromatin is comprised of genomic DNA wrapped around a core of four histone proteins that include histone 2A, 2B, 3 and 4 (Strahl & Allis, 2000). The spatial relationship between DNA and the histone core determines the transcriptional status of surrounding genes. Where DNA is tightly associated with histones, the transcriptional initiation complex (TIC) is excluded from the region, resulting in transcriptional repression (McKinsey et al. 2001). Conversely, a loose association between DNA and histones allows the TIC complex access to promoter regions, resulting in transcriptional activation (McKinsey et al. 2001). The spatial association between DNA and histones is largely governed by post-translational modifications to histone proteins, particularly histones 3 and 4 (Strahl & Allis, 2000). These modifications, or epigenetic marks, include phosphorylation, acetylation and mono-, di- and tri-methylation (Kouzarides, 2007). While some of these modifications are associated with transcriptional repression, acetylation of lysine residues within histone 3 correlates with transcriptional activation (Kouzarides, 2007). Histone acetylation creates an open chromatin confirmation by neutralising the positively charged lysine side chains, breaking the tight electrostatic interaction between the negatively charged DNA phosphate backbone (Strahl & Allis, 2000). Specifically, histone 3, lysine 9 and 14 (H3K9/14) and H3K36 acetylation are associated with transcriptional initiation and elongation, respectively (Kuo et al. 1996; Morris et al. 2007). While it is clear that exercise induces the expression of a network of skeletal muscle genes, it is unclear if these mechanisms play a role in this response.
Histone acetylation is regulated by a balance between histone acetyl-transferase (HAT) and histone deacetylase (HDAC) activities (McKinsey et al. 2001). Rodent studies have shown that the class IIa HDAC enzymes – consisting of isoforms 4, 5, 7 and 9 – play a key role in skeletal muscle development and phenotype (Potthoff et al. 2007). These enzymes do not possess HDAC activity themselves, but rather recruit a repressive complex containing HDAC3 for this purpose (Fischle et al. 2002). They are regulated by phosphorylation-dependent nuclear export (McKinsey et al. 2000) and ubiquitin-mediated proteasomal degradation (Potthoff et al. 2007), both of which remove their repressive function. The class IIa HDAC family redundantly regulate skeletal muscle fibre type and oxidative metabolism. In addition, a recent study has shown that overexpression of HDAC5 in mouse skeletal muscle is sufficient to attenuate adaptations to exercise training (Potthoff et al. 2007). This fits with our previous observation that the nuclear abundance of HDAC5 is reduced in human skeletal muscle following a single bout of exercise (McGee & Hargreaves, 2004) and suggests that the class IIa HDACs and chromatin remodelling could be critical regulators of exercise adaptations. Therefore, the purpose of this study was to examine whether exercise results in histone modifications associated with chromatin remodelling and transcriptional activation in human skeletal muscle, and to determine whether this is associated with regulation of the class IIa HDAC enzymes.
Methods
Subjects
Nine male subjects (23 ± 1 years: 75 ± 6 kg; 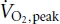 = 41 ± 3 ml kg−1 min−1) were recruited for the study after completing a medical questionnaire and giving their informed, written consent. All experimental procedures were approved by The University of Melbourne Human Research Ethics Committee in accordance with the declaration of Helsinki. At least 7 days prior to the experimental trial, all subjects performed an incremental cycling (Lode, Groningen, The Netherlands) test to fatigue to determine peak pulmonary oxygen uptake (
= 41 ± 3 ml kg−1 min−1) were recruited for the study after completing a medical questionnaire and giving their informed, written consent. All experimental procedures were approved by The University of Melbourne Human Research Ethics Committee in accordance with the declaration of Helsinki. At least 7 days prior to the experimental trial, all subjects performed an incremental cycling (Lode, Groningen, The Netherlands) test to fatigue to determine peak pulmonary oxygen uptake (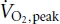 ). This test was also used to select the power output for the experimental trial from the linear relationship between oxygen uptake and power output.
). This test was also used to select the power output for the experimental trial from the linear relationship between oxygen uptake and power output.
Exercise
Subjects performed a single bout of cycling for 60 min at 76 ± 2% of 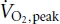 , after a 12 h, overnight fast. Expired air was collected twice, between 15–20 and 40–45 min to ensure that subjects were exercising at the expected exercise intensity.
, after a 12 h, overnight fast. Expired air was collected twice, between 15–20 and 40–45 min to ensure that subjects were exercising at the expected exercise intensity.
Muscle biopsies
Muscle samples were obtained from the vastus lateralis prior to and immediately following exercise using the percutaneous needle biopsy technique with suction. Muscle samples were immediately frozen in liquid nitrogen and stored for later analysis.
Protein extraction
Nuclear proteins were isolated as previously described (McGee et al. 2003), with the addition of 10 μm 3,5-bis-(4-boronic acid-benzylidene)-1-methylpiperidin-4-one (Calbiochem, San Diego, CA, USA) to all buffers. For whole muscle proteins, approximately 15 mg of muscle were homogenised in 10 volumes of homogenisation buffer (50 mm Tris pH 7.5, 1 mm EDTA, 1 mm EGTA, 10% glycerol, 1% Triton X-100, 50 mm NaF, 5 mm sodium pyrophosphate, 1 mm DTT, 1 μl (5 mg tissue)−1 protease inhibitor cocktail (Sigma, St Louis, MD, USA) 10 μm 3,5-bis-(4-boronic acid-benzylidene)-1-methylpiperidin-4-one) for approximately 30 s on ice. The homogenate was then spun in a centrifuge for 5 min at 1000 g at 4°C. The supernatant was extracted and stored for later analysis. Protein concentration was determined using the bicinchoninic acid (BCA) method.
Immunoprecipitation
Whole muscle protein (500 μg) was made up to 500 μl in homogenisation buffer. Samples were incubated with 4 μg of anti-ubiquitin (Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibody overnight at 4°C. Samples were incubated with 50 μl of 50% Protein A sepharose bead slurry (Amersham Biosciences, Castle Hill, Sydney, Australia) for 2 h, while rotating at 4°C. The sepharose-bound immune complex was pelleted by centrifugation and washed 4 times with 1 ml of 1 × phosphate-buffered saline. Immune complexes were then resolved by SDS-PAGE and the amounts of HDAC4, 5, 7 and 9 were determined by probing with antibodies directed towards those proteins as described below. A control lane of beads only was included on all gels to ensure that the signals observed were specific to the immune complex, and not the beads themselves.
Immunoblotting
Proteins were separated and identified using SDS-PAGE. Twenty-five micrograms of protein from the whole cell homogenate and 40 μg of nuclear protein from each sample were loaded onto 1.5 mm 8% acrylamide gels before undergoing electrophoresis for 50 min at 180 V. Proteins were transferred to a nitrocellulose membrane for 100 min at 100 V. Membranes were blocked for 1 h in blocking buffer (5% skim milk powder or 5% bovine serum albumin in Tris-buffered saline and 0.25% Tween (TBST)) and exposed overnight, at 4°C, to primary antibodies for HDAC4, 5, 7 (Cell Signalling Technology, Beverly, MA, USA) and 9 (Santa Cruz Biotechnology), phosphorylated (T172) AMPK α, phosphorylated (S744/748) PKD, phosphorylated (T287) CaMKII (Cell Signalling Technology) and total and acetylated (K9/14 and K36) H3 (Millipore, Billerica, MA, USA). Membranes were exposed to appropriate anti-species horseradish peroxidase (HRP) conjugated secondary antibodies, at a concentration of 1 in 10 000 in 1 × TBST, for 60 min at room temperature. Membranes were incubated in enhanced chemiluminescence substrate (ECL Plus WB Detection Reagent, GE Healthcare, Uppsala, Sweden) and bands visualized using a Bio-Rad Chemi-Doc System (Bio-Rad, Hercules, CA, USA). Bands were quantified using Bio-Rad Quantity One 1D Image Analysis Software.
Real-time RT PCR
Total RNA was extracted from approximately 10 mg of muscle using the Aurum total RNA Fatty and Fibrous tissue kit (BioRad). RNA was reverse transcribed to cDNA using the iScript kit (BioRad). Forward and reverse primers complimentary to the human HDAC4, 5, 7 and 9 were designed by using OligoPerfect software (Invitrogen). Primer sequences are available upon request. Real-time PCR was performed using an iCycler thermal cycler with IQ5 detection system and software with Sybr green chemistry (BioRad). Gene expression values were normalized to the housekeeping gene cyclophilin.
HDAC activity
HDAC activity was determined using a microplate colorimetric kit (Biomol, Matford Court, UK), according to manufacturer's instructions. Briefly, 10 μg of whole muscle protein in 5 μl were added to assay buffer and HDAC substrate to a total volume of 50 μl. Following addition of 50 μl developer solution, samples were incubated at 37°C for 15 min. Sample absorbance was measured at 405 nm. Negative controls where the HDAC inhibitor TSA was also included in the reaction mix were also performed. Addition of TSA inhibited all HDAC activity (data not shown).
Statistical analyses
All values reported are means ± standard error of the mean (s.e.m.), with resting samples assigned the arbitrary value of 1.0 and post-exercise samples expressed relative to rest. Resting and post-exercise means were compared using a t test with a significance level of 0.05.
Results
Exercise induces alterations in histone modifications associated with transcriptional activation
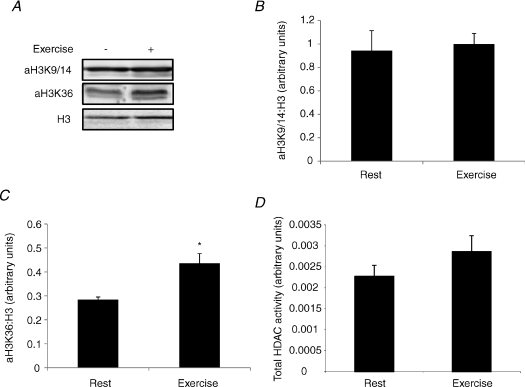
Exercise is associated with the enhanced expression of a number of genes (Mahoney et al. 2005). To determine whether exercise results in histone modifications associated with transcriptional activation, we analysed muscle samples for global H3 acetylation (Fig. 1A). Immediately after exercise, H3K9/14 acetylation was not significantly different from pre-exercise values (Fig. 1B). However, H3K36 acetylation, which is thought to regulate transcriptional elongation, was increased by 64% (P < 0.05) after exercise when compared with rest (Fig. 1C), suggesting that exercise induces chromatin remodelling associated with enhanced transcription.
Figure 1. Exercise induces chromatin remodelling.
A, representative immunoblots of H3K9/14 and H3K36 acetylation and total H3 levels before and immediately after exercise. B, the H3K9/14 to total H3 ratio before and immediately after exercise. C, the H3K36 to total H3 ratio before and immediately after exercise. D, total HDAC activity before and immediately after exercise. All values are reported as means ±s.e.m. (n= 6–9). *Significantly different from rest (P= 0.01).
Exercise does not alter global HDAC activity
Histone acetylation levels reflect the balance between HAT and HDAC activities (McKinsey et al. 2001) and overexpression of HDAC5 is sufficient to block skeletal muscle adaptations to exercise training (Potthoff et al. 2007). Consequently, we assessed whether the increase in H3K36 acetylation following exercise was associated with a decrease in global HDAC activity. However, we found no statistical difference (P= 0.31) in skeletal muscle HDAC activity immediately following exercise (Fig. 1D).
Exercise alters class IIa HDAC function
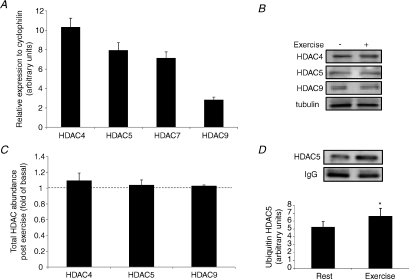
Although we observed no difference in total HDAC activity following exercise, regulation of the class IIa HDACs will not necessarily be reflected by changes in this parameter, as they do not possess intrinsic HDAC activity (Fischle et al. 2002). Alterations in class IIa HDAC function can occur through proteasomal degradation (Potthoff et al. 2007) and changes in subcellular localisation (McKinsey et al. 2000). A recent study has found that ubiquitin-mediated proteasomal degradation of class IIa HDACs contributes to skeletal muscle adaptations in response to exercise training (Potthoff et al. 2007). To assess whether these HDACs are involved in exercise adaptations in human skeletal muscle, we first determined whether the class IIa family are expressed in this tissue. Using RT-PCR, HDAC4, 5, 7 and 9 were all found to be expressed in human skeletal muscle (Fig. 2A). To determine whether proteasomal degradation of class IIa HDACs occurs during exercise, we analysed total protein levels of HDAC4, 5, 7 and 9 immediately after exercise (Fig. 2B). We were unable to reliably detect HDAC7 in our samples. There was no difference in the abundance of HDAC4, 5 and 9 immediately following exercise (Fig. 2C), suggesting that proteasomal degradation does not play a role in chromatin remodelling during exercise. This does not rule out this mechanism regulating exercise adaptations in the post-exercise period. To gain an insight into whether ubiquitin-mediated proteasomal degradation might play a role in the post-exercise period, we also analysed class IIa HDAC ubiquitination immediately following exercise. Using coimmunoprecipitation techniques, we found that HDAC5 ubiquitination was increased by ∼30% (P < 0.05) immediately following exercise (Fig. 2D). We could not detect ubiquitination of any other class IIa HDAC.
Figure 2. Class IIa HDAC expression in human skeletal muscle and following exercise.
A, class IIa HDAC mRNA levels expressed relative to cyclophilin. B, representative immunoblots of total class IIa HDAC protein before and immediately after exercise. C, total class IIa HDAC protein immediately after exercise expressed relative to resting levels. D, ubiquitin-associated HDAC5 before and immediately after exercise. All values are reported as means ±s.e.m. (n= 6–9). *Significantly different from rest (P= 0.04).
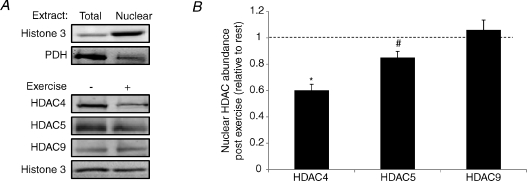
Phosphorylation-dependent nuclear export of class IIa HDACs also plays a critical role in their regulation (McKinsey et al. 2000). We analysed the nuclear abundance of the class IIa HDACs immediately following exercise (Fig. 3A). HDAC4 and 5 levels in the nucleus were significantly decreased by 40% (P < 0.05) and 16% (P < 0.05), respectively, immediately after exercise (Fig. 3B). As total levels of these HDACs did not change, this suggests nuclear export of HDAC4 and 5. There was no difference in the nuclear abundance of HDAC9 (Fig. 3B). These data suggest that phosphorylation-dependent nuclear export of HDAC4 and 5 could play a role in chromatin remodelling during exercise.
Figure 3. Exercise induces the nuclear export of HDAC4 and 5.
A, representative immunoblots confirming nuclear enrichment and the nuclear abundance of class IIa HDAC protein before and immediately after exercise. B, nuclear class IIa HDAC abundance immediately after exercise expressed relative to resting levels. All values are reported as means ±s.e.m. (n= 8–9). *Significantly different from rest (P < 0.01). #Significantly different from rest (P= 0.03).
Exercise activates multiple class IIa HDAC kinases
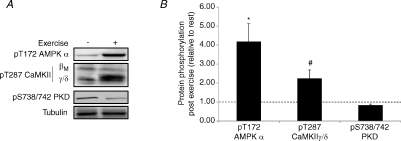
A number of kinases whose activities are modulated by factors associated with exercise are capable of phosphorylating the class IIa HDACs and inducing their nuclear export. This includes the AMP-activated protein kinase (AMPK; McGee et al. 2008), the calcium–calmodulin-dependent protein kinase II (CaMKII; Backs et al. 2008) and protein kinase D (PKD; Chang et al. 2005). We analysed the phosphorylation status of these kinases (Fig. 4A), which is a proxy measure of their activation. Immediately after exercise, AMPK and CaMKII phosphorylation was increased to ∼4.1-fold (P < 0.05) and ∼2.2-fold (P < 0.05) above resting levels, respectively (Fig. 4B). PKD phosphorylation was 17% lower than basal levels immediately after exercise, although this was not statistically significant (P= 0.06; Fig. 4B).
Figure 4. Exercise activates kinases that mediate phosphorylation dependent nuclear export of HDAC4 and 5.
A, representative immunoblots of pT172 AMPK α, pT287 CaMKII βM and γ/δ and pS738/742 PKD before and immediately after exercise. B, pT172 AMPK α, pT287 CaMKII γ/δ and pS738/742 PKD phosphorylation immediately after exercise expressed relative to resting levels. All values are reported as means ±s.e.m. (n= 7–9). *Significantly different from rest (P < 0.01). #Significantly different from rest (P < 0.01).
Discussion
Although it has been known for some time that exercise enhances the expression of a subset of genes involved in skeletal muscle phenotypic adaptation, the molecular mechanisms responsible remain to be fully determined. Chromatin remodelling has emerged as one of the major mechanisms controlling gene expression (Strahl & Allis, 2000). Histone modifications regulate myosin heavy chain isoform expression in response to muscle unloading (Pandorf et al. 2009). However, to date, no studies have examined the effects of exercise on global histone modifications mediating chromatin remodelling. Here we showed that immediately after exercise in human skeletal muscle, H3K36 acetylation is significantly increased. This modification is associated with promoter regions that contain active RNA polymerase II (RNAPII; Morris et al. 2007). This finding was also associated with the nuclear export of the class IIa HDAC isoforms 4 and 5, which are members of a group of enzymes that control histone acetylation levels (McKinsey et al. 2001). In addition, we show activation of AMPK and CaMKII, which are two kinases that can induce nuclear export of HDAC4 and 5 (Back et al. 2008; McGee et al. 2008). These data delineate a signalling pathway that might mediate gene transcription in human skeletal muscle in response to exercise.
Although we saw an increase in the global abundance of H3K36 acetylation immediately following exercise, we did not observe any change in H3K9/14 acetylation. As the latter modification is thought to be a prelude to transcriptional initiation (Nowark & Corces, 2004), these data can be interpreted in a number of different ways. The first interpretation is that H3K9/14 acetylation is not required to initiate transcription during exercise. Indeed, it has been found that RNAPII occupies inactive promoter regions and that these genes are regulated at the transition between transcriptional initiation and elongation (Hargreaves et al. 2009). It could be that genes that activated during exercise already have RNAPII situated at their promoter region, and only require H3K36 acetylation to initiate transcriptional elongation. This paradigm has been observed for primary response genes that are immediately activated following lipopolysaccharide exposure in macrophages (Hargreaves et al. 2009). An alternative interpretation is that there is a temporal dependency of H3K9/14 acetylation in response to exercise. It is possible that H3K9/14 acetylation is increased early in the exercise stimulus to initiate the transcription of genes that are immediately up-regulated in response to exercise. Another alternate explanation is that global H3K9/14 acetylation might not be representative of acetylation patterns at individual promoter regions that are regulated by exercise. Indeed, chromatin immunoprecipitation assays with an antibody directed towards this modification coupled with promoter array technology or next-generation sequencing could unveil the spatial and temporal changes in promoter H3K9/14 acetylation during exercise.
Despite observing an increase in H3K36 acetylation following exercise, we did not observe a concomitant decrease in HDAC activity. This is not entirely surprising when you consider the function of the class IIa HDACs, which has been implicated in the adaptive response to exercise (Potthoff et al. 2007). These transcriptional repressors do not contain any intrinsic HDAC activity themselves, but instead recruit a complex containing HDAC3 to gain this activity (Fischle et al. 2002). In this sense, the class IIa HDACs merely act as scaffolds that engage HDAC activity to a particular promoter region. Hence, regulation of the class IIa HDACs, even through degradation by the proteasome, should not have any effect on total HDAC activity. Instead, regulation of the class IIa HDACs will relocalise HDAC activity away from the promoter regions that they regulate.
A previous study has implicated ubiquitin-mediated proteasomal degradation in the adaptive response of skeletal muscle to exercise (Potthoff et al. 2007). This study examined myocyte enhancer factor 2 (MEF2)-driven lacZ expression in the skeletal muscle of mice after 3 h of forced treadmill exercise following acute treatment with the proteasome inhibitor MG132, or vehicle control. LacZ expression was significantly reduced in the skeletal muscles of MG132-treated mice (Potthoff et al. 2007). The authors subsequently showed that HDAC protein abundance was higher in mice treated with MG132. We have examined this protocol in human skeletal muscle in the present study following 60 min of cycling, by assessing the total protein abundance of HDAC4, 5 and 9 immediately after exercise. We saw no change in HDAC abundance immediately post-exercise, suggesting that HDAC proteasomal degradation during exercise is not a major contributing factor to the skeletal muscle adaptive response. However, it is possible that this mechanism becomes more important in prolonged bouts of exercise. It should be noted that the effect of MG132 in the Potthoff et al. (2007) study was examined after 3 h of treadmill running. Indeed, we did observe an increase in ubiquitin-associated HDAC5 immediately after exercise, which could also suggest that proteasomal degradation of HDACs might play a role in the post-exercise period.
In contrast to ubiquitin-mediated proteasomal degradation of class IIa HDACs, we did find that exercise resulted in the nuclear export of HDAC4 and 5. These findings agree with our previous finding that HDAC5 is exported from the nucleus in human skeletal muscle during exercise (McGee & Hargreaves 2004). These data suggest that phosphorylation-dependent nuclear export of class IIa HDACs is the preferred regulatory mechanism in response to acute metabolic stress. It is interesting that HDAC4 and 5 were the two isoforms found to undergo nuclear export in response to exercise. It has recently been found that HDAC4 and 5 preferentially form oligomers, which influences their sensitivity to upstream signalling (Backs et al. 2008). For example, HDAC4 has a domain unique from other class IIa HDACs located within the core of its sequence that interacts with CaMKII (Backs et al. 2008). Phosphorylation of HDAC4 by CaMKII and its subsequent nuclear export can in turn export HDAC5 from the nucleus when these two class IIa HDACs are associated (Backs et al. 2008). Thus, although HDAC5 cannot be directly phosphorylated by CaMKII, it can still be exported from the nucleus in response to CaMKII signalling. Indeed, the importance of the CaMK family for histone acetylation at the GLUT4 gene during exercise (Smith et al. 2008) and in response to calcium perturbations (Mukwevho et al. 2008) has been observed previously.
Despite CaMKII being a unique kinase for HDAC4 within the class IIa HDAC family, a number of other kinases are known to phosphorylate the class IIa HDACs. These include AMPK (McGee et al. 2008), PKD and members of the AMPK-related kinases, such as Mark2 (Chang et al. 2005). However, Mark2 and many of the other AMPK-related kinases are not activated by muscle contraction (Sakamoto et al. 2004). This led us to examine the activation state of CaMKII, AMPK and PKD in the present study using phospho-specific antibodies. We found significant increases in AMPK and CaMKII activation, which is consistent with previous research (Hutber et al. 1997; Rose & Hargreaves, 2003). However, PKD phosphorylation tended to decrease with exercise, although this was not statistically significant (P= 0.06). This was particularly surprising given that PKD is sensitive to metabolic perturbations such as increases in oxidative stress and diacylglycerol concentrations (Rozengurt et al. 2005). Indeed, PKD has been proposed as a contraction-sensitive kinase in the heart (Luiken et al. 2008). These data suggest that AMPK and CaMKII contribute to the nuclear export of HDAC4 and 5, thereby possibly contributing to the associated increase in H3K36 acetylation. Indeed, it is conceivable that redundancy between AMPK and CaMKII signalling to the class IIa HDACs exists. A recent paper has examined this in the context of regulation of the GLUT4 gene (Murgia et al. 2009), whose expression is increased in human skeletal muscle in response to exercise (Kraniou et al. 2000). This study (Murgia et al. 2009) found that genetic inhibition of both AMPK and CaMKII, but not these pathways exclusively, was required to reduce GLUT4 expression. These data and that of the present study suggest that AMPK and CaMKII might be equally important in mediating skeletal muscle adaptations to exercise.
A number of additional points need to be considered when interpreting data from the present study. Potthoff et al. (2007) have established that proteasomal degradation of the class IIa HDACs occurs in a fibre-type-dependent manner. We obtained samples of the mixed vastus lateralis muscle obtained by percutaneous needle biopsy from different sites before and after exercise. This means that the fibre-type distribution between pre- and post-exercise samples could be slightly different and adds a confounding variable to our data. However, given the small degree of variability in our total class IIa HDAC protein measurements, we feel we have a similar fibre-type distribution in our pre- and post-exercise muscle samples. Finally, we did not control deubiquitinating activity in our muscle lysates and this might have contributed to the fact that we were unable to detect HDAC4 and 9 in an ubiquitin immune complex.
In conclusion, we have showed that exercise results in a global increase in H3K36 acetylation, a histone modification that is associated with transcriptional elongation. This finding was also associated with the nuclear export of HDAC4 and 5, rendering these enzymes unable to suppress histone acetylation. Finally, these data were associated with activation of AMPK and CaMKII, two kinases that are able to induce nuclear export of HDAC4 and 5. Together, these findings delineate a signalling pathway that might mediate skeletal muscle adaptations to exercise.
Acknowledgments
We wish to thank the subjects for their participation in the study. S.L.M. is a NHMRC Peter Doherty Fellow (400446).
Glossary
Abbreviations
- AMPK
AMP-activated protein kinase
- CaMKII
calcium–calmodulin-dependent protein kinase II
- H3K9/14
histone 3 lysine 9 and 14
- H3K36
histone 3 lysine 36
- HAT
histone acetyl-transferase
- HDAC
histone deacetylase
- GLUT4
glucose transporter isoform 4
- PKD
protein kinase D
- RNAPII
RNA polymerase II
- TIC
transcriptional initiation complex
Author contributions
All authors planned and undertook the experiments and contributed to data analysis and writing of the paper.
References
- Backs J, Backs T, Bezprozvannaya S, McKinsey TA, Olson EN. Histone deacetylase 5 acquires calcium/calmodulin-dependent kinase II responsiveness by oligomerization with histone deacetylase 4. Mol Cell Biol. 2008;28:3437–3445. doi: 10.1128/MCB.01611-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Bezprozvannaya S, Li S, Olson EN. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc Natl Acad Sci U S A. 2005;102:8120–8125. doi: 10.1073/pnas.0503275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutber CA, Hardie DG, Winder WW. Electrical stimulation inactivates muscle acetyl-CoA carboxylase and increases AMP-activated protein kinase. Am J Physiol Endocrinol Metab. 1997;272:E262–E266. doi: 10.1152/ajpendo.1997.272.2.E262. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their functions. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kraniou Y, Cameron-Smith D, Misso M, Collier G, Hargreaves M. Effects of exercise on GLUT-4 and glycogenin gene expression in human skeletal muscle. J Appl Physiol. 2000;88:794–796. doi: 10.1152/jappl.2000.88.2.794. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- Luiken JJ, Vertommen D, Coort SL, Habets DD, El Hasnaoui M, Pelsers MM, Viollet B, Bonen A, Hue L, Rider MH, Glatz JF. Identification of protein kinase D as a novel contraction-activated kinase linked to GLUT4-mediated glucose uptake, independent of AMPK. Cell Signal. 2008;20:543–556. doi: 10.1016/j.cellsig.2007.11.007. [DOI] [PubMed] [Google Scholar]
- McGee SL, Hargreaves M. Exercise and myocyte enhancer factor 2 regulation in human skeletal muscle. Diabetes. 2004;53:1208–1214. doi: 10.2337/diabetes.53.5.1208. [DOI] [PubMed] [Google Scholar]
- McGee SL, Howlett KF, Starkie RL, Cameron-Smith D, Kemp BE, Hargreaves M. Exercise increases nuclear AMPK a2 in human skeletal muscle. Diabetes. 2003;52:926–928. doi: 10.2337/diabetes.52.4.926. [DOI] [PubMed] [Google Scholar]
- McGee SL, van Denderen BJ, Howlett KF, Mollica J, Schertzer JD, Kemp BE, Hargreaves M. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- Mahoney DJ, Parise G, Melov S, Safdar A, Tarnopolsky MA. Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. FASEB J. 2005;19:1498–1500. doi: 10.1096/fj.04-3149fje. [DOI] [PubMed] [Google Scholar]
- Morris SA, Rao B, Garcia BA, Hake SB, Diaz RL, Shabanowitz J, Hunt DF, Allis CD, Lieb JD, Strahl BD. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J Biol Chem. 2007;282:7632–7640. doi: 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukwevho E, Kohn TA, Lang D, Nyatia E, Smith J, Ojuka EO. Caffeine induces hyperacetylation of histones at the MEF2 site on the Glut4 promoter and increases MEF2A binding to the site via a CaMK-dependent mechanism. Am J Physiol Endocrinol Metab. 2008;294:E582–E588. doi: 10.1152/ajpendo.00312.2007. [DOI] [PubMed] [Google Scholar]
- Murgia M, Elbenhardt Jensen T, Cusinato M, Garcia M, Richter EA, Schiaffino S. Multiple signalling pathways redundantly control GLUT4 gene transcription in skeletal muscle. J Physiol. 2009;587:4319–4327. doi: 10.1113/jphysiol.2009.174888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowark SJ, Corces VG. Phosphorylation of histone 3: a balancing act between chromosome condensation and transcriptional activation. Trends Gen. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Pandorf CE, Haddad F, Wright C, Bodell PW, Baldwin KM. Differential epigenetic modifications of histones at the myosin heavy chain genes in fast and slow skeletal muscle fibres and in response to muscle unloading. Am J Physiol Cell Physiol. 2009;297:C6–C16. doi: 10.1152/ajpcell.00075.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Wu H, Arnold MA, Shelton JM, Backs J, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibres. J Clin Invest. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AJ, Hargreaves M. Exercise increases Ca2+–calmodulin-dependent protein kinase II activity in human skeletal muscle. J Physiol. 2003;553:303–309. doi: 10.1113/jphysiol.2003.054171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozengurt E, Rey O, Waldron RT. Protein kinase D signalling. J Biol Chem. 2005;280:13205–13208. doi: 10.1074/jbc.R500002200. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Göransson O, Hardie DG, Alessi DR. Activity of LKB1 and AMPK-related kinases in skeletal muscle: effects of contraction, phenformin, and AICAR. Am J Physiol Endocrinol Metab. 2004;287:E310–E317. doi: 10.1152/ajpendo.00074.2004. [DOI] [PubMed] [Google Scholar]
- Smith JA, Collins M, Grobler LA, Magee CJ, Ojuka EO. Exercise and CaMK activation both increase the binding of MEF2A to the Glut4 promoter in skeletal muscle in vivo. Am J Physiol Endocrinol Metab. 2008;292:E413–E420. doi: 10.1152/ajpendo.00142.2006. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]