Abstract
Much evidence strongly suggests a positive role for one or more E2F species in the control of exit from G0/G1. Results described here provide direct evidence that endogenous E2F-1, as predicted, contributes to progression from G0 to S. By contrast, cycling cells lacking an intact E2F-1 gene demonstrated normal cell cycle distribution. Therefore, E2F-1 exerts a unique function leading to timely G0 exit of resting cultured primary cells, while at the same time being unnecessary for normal G1 to S phase progression of cycling cells.
Keywords: RB, cell cycle control
The retinoblastoma tumor susceptibility gene product, pRB, has been implicated in the control of various cellular processes. They include entry into S phase, the activity of certain transcription factors and enzymes, maintenance of certain cellular differentiation events, and programmed cell death. A widely held view suggests that pRB functions in cell cycle control, in part, by binding to and modulating the transcription regulation behavior of a major cellular partner, E2F-1. E2F-1 is a member of a transcription factor family that consists of six E2F species (E2F-1, -2, -3, -4, -5, and -6) and two DP members (DP-1 and -2) (for reviews and references see refs. 1–6). E2Fs and DPs form heterodimers and are responsible for the transcription of several genes that are important for cell cycle progression (7–10). The transcription activity of E2F-1 is tightly regulated through the cell cycle. In G1, hypophosphorylated pRB binds to E2F-1 suppressing the transactivation potential of the protein and converting it into a repressor (11–15). In late G1, pRB is phosphorylated by cyclin-dependent kinases, leading to the release of E2F-1, which in turn acquires an ability to activate certain promoters (1, 16–19). After peaking at G1/S and during early S phase, the transactivation activity of free E2F-1 decreases in mid-late S phase (10, 16). The down regulation of E2F-1 transactivation function is likely, in part, because of the rapid degradation of free E2F-1 through the ubiquitin–proteasome pathway (20–22). In addition, in late G1 and S, E2F-1 binds cyclin A kinase. The bound enzyme phosphorylates a DP species heterodimerized to E2F-1, resulting in loss of E2F-1/DP DNA binding activity (23–26).
Overproduction of E2F-1 can lead to neoplastic transformation of some immortalized cell lines, and deregulated E2F-1 synthesis can drive serum-starved fibroblasts into S phase (for review see ref. 1). This suggests that E2F-1 has the potential to drive G0 cells into the cell cycle. On the other hand, recent evidence from the Nevins laboratory shows that, unlike E2F-3, E2F-1 is not essential for G1 exit of proliferating cells (10). This notwithstanding, whether or not endogenous E2F-1 participates in the G0 exit process has not been determined. In this report we describe the results of experiments on genetically defined, primary mouse embryo fibroblasts aimed at determining whether endogenous E2F-1 plays a discrete role in the cycling of proliferating cells and/or in G0 exit regulation. In keeping with the results of Leone et al. (10), the data suggest that E2F-1 is not required for G1 exit of cycling cells. By contrast, it plays an important role in regulating the emergence of G0-arrested cells into the cell cycle.
MATERIALS AND METHODS
Cells and Cell Cycle Analysis.
Cells were cultivated in DMEM containing 10% fetal calf serum in a 10% CO2-containing atmosphere. They were detached from plates by trypsinization. After washing twice with PBS buffer containing 10 mM Hepes (pH 7.3) and 0.1% BSA, cells were fixed in 70% ethanol at 4°C. Before cell cycle analysis, cells were washed with PBS containing 10 mM Hepes (pH 7.3), then resuspended in PI buffer (38 mM sodium citrate/69 μM propidium iodide/5 μg/ml RNase), and incubated at 37°C for 30 min. Cell cycle profiles were analyzed by fluorescence-activated cell sorter (FACS) in a Becton Dickinson FACScan machine using the cellfit program.
Western Blot Analysis.
Proteins were resolved by SDS/PAGE, transferred to nitrocellulose membranes (Schleicher & Schuell), and immunoblotted with rabbit polyclonal anti-cyclin E Ab (M20; Santa Cruz Biotechnology). Antibody detection was achieved upon incubation with horseradish peroxidase-conjugated donkey anti-rabbit IgG (Amersham), followed by enhanced chemiluminescence (Amersham). These procedures were performed according to the manufacturer’s protocol.
Mouse Embryo Fibroblast (MEF) Preparation.
MEFs were prepared from individual embryos at embryonic day 14.5 (E14.5; 129/Sv) bearing E2F-1+/+, E2F-1+/−, and E2F-1−/− genotypes. The head and internal organs were removed, and the torso was minced and dispersed in 0.1% trypsin (45–60 min at 37°C). Cells were grown for two population doublings (considered as one passage) and then viably frozen. These MEFs were used for all subsequent experiments. MEFs were maintained in DMEM containing 10% fetal bovine serum (FBS; GIBCO) and subcultured 1:4 upon reaching confluence. For serum starvation experiments, MEFs were plated in DMEM containing 0.1% FBS and then incubated at 37°C for 72 hr, before stimulation with DMEM containing 10% FBS.
RESULTS
S Phase Entry Is Delayed in E2F-1−/− MEFs.
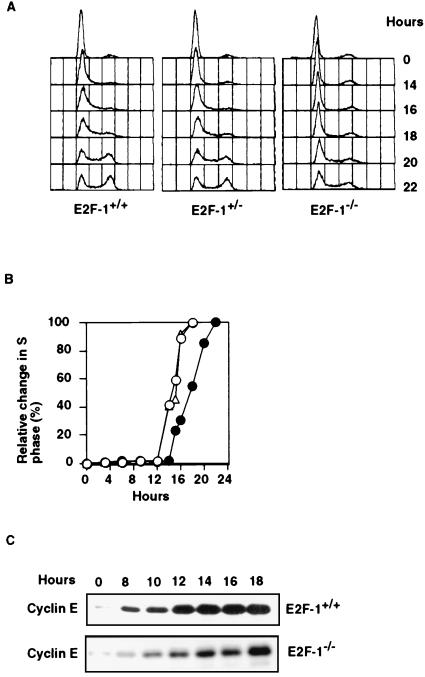
E2F-1, when overproduced, can activate several genes necessary for S phase progression. However, the physiological role of endogenous E2F-1 in cell cycle regulation is still unclear. To determine whether E2F-1 function is necessary for G1 exit of cycling diploid cells, we analyzed the behavior of MEFs isolated from E14.5 E2F-1+/+, E2F-1+/−, and E2F-1−/− embryos (27). After serum starvation for 72 hr, cells were released from G0 by addition of 10% serum-containing medium and collected at specific intervals thereafter. They were then subjected to FACS analysis, the results of which are shown in Fig. 1A. E2F-1−/− cells exhibited prolonged G0 to S phase transit, emerging 3–4 hr later than their E2F-1+/+ or E2F-1+/− counterparts, which were cultivated, harvested, and analyzed in parallel (Fig. 1 A and B). By contrast, no significant differences were noted in cell cycle intervals among these same MEF strains when all were continuously cultivated in serum-containing medium (Table 1). This is in keeping with previously published results (10, 27, 42). Therefore, it would appear that there is an E2F-1-dependent proliferation effect in cells emerging from G0 rather than in cycling cells.
Figure 1.
Cell cycle kinetics of E2F-1 MEFs. (A) Cell cycle profiles of serum-starved E2F-1+/+, E2F-1+/−, and E2F-1−/− MEFs at indicated times after readdition of serum. (B) Relative increases in S phase E2F-1+/+ (○), E2F-1+/− (Δ), and E2F-1−/− (•) MEFs. (C) Western blot analysis of cyclin E in serum-starved E2F-1+/+ and E2F-1−/− MEFs at various times after serum stimulation.
Table 1.
Cell cycle distribution of asynchronously growing MEF cells of various E2F-1 genotypes
| E2F-1 | G1 | S | G2/M |
|---|---|---|---|
| +/+ | 46.3% | 34.3% | 19.4% |
| +/− | 46.4% | 33.2% | 20.4% |
| −/− | 47.5% | 34.5% | 18.0% |
In an analogous experiment, we determined the relative levels of cyclin E in E2F-1+/+ and E2F-1−/− cells at various times after exit from G0. Cyclin E is a pivotal controller of events in mid-to-late G1 that licenses S phase entry (28–30). As shown in Fig. 1C, the induction of cyclin E protein levels in E2F-1−/− cells was also delayed by 3–4 hr compared with the timing in E2F-1+/+ cells. Hence, there is an apparent correlation between timely E2F-1 synthesis and timely synthesis of a primary effector of G1 exit.
The Restriction Point Is Maintained in E2F-1−/− MEFs.
The restriction (R) point is believed to be a critical checkpoint affecting G1 progression (31). By definition, once a cell passes the R point, it progresses into S phase in a serum-independent and cycloheximide-resistant manner. In short, it no longer requires exogenous growth factors to progress through the cycle, nor does it require the synthesis of new proteins. It has been shown that the R point transition and the onset of RB phosphorylation occur almost simultaneously in mid/late G1 phase, and the R point control is disrupted in RB−/− MEF cells (32–36). Given that the phosphorylation and inactivation of pRB lead to the dissociation of E2F-1 from an RB-E2F-1/DP complex, we asked whether the R point is maintained in E2F-1−/− cells.
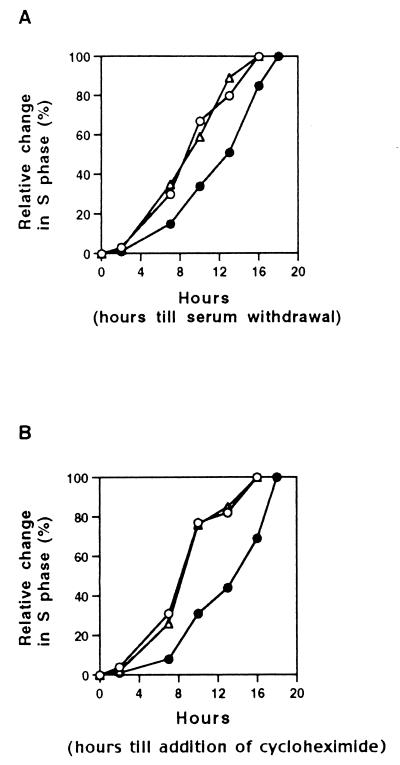
E2F-1 MEFs were growth-arrested by serum starvation and then allowed to reenter the cell cycle after serum addition. At various intervals thereafter, serum was either removed or cycloheximide (final concentration = 50 ng/ml) was added to the medium. The fraction of cells entering S phase was then analyzed by FACS at defined time points. As shown in Fig. 2, all three MEF strains displayed increasing resistance to both serum withdrawal and cycloheximide, which began to appear at approximately the same time, i.e., about 4–6 hr after serum refeeding. (Fig. 2). This indicates that the restriction point point was maintained in both mutant cell strains and was detected at the same time in all three cultures. However, after the restriction point, the E2F-1−/− cells progressed more slowly into S phase, in keeping with the results shown in Fig. 1. These data indicate that loss of E2F-1 function does not affect the ability of a primary mouse embryo fibroblast to become growth factor-independent after a specific point during G0 exit. Rather, it affects events after the restriction point, in keeping with the delayed onset of cyclin E observed in nullizygous cells (Fig. 1).
Figure 2.
Analysis of the restriction point in E2F-1−/− MEFs. E2F-1+/+ (○), E2F-1+/− (Δ), and E2F-1−/− (•) MEFs were serum-starved for 72 hr, and then serum was added to the medium. After the indicated periods of serum stimulation, cells were again deprived of serum (A) and subsequently analyzed for cell cycle progression. Alternatively, cycloheximide was added to the medium (final concentration of 50 ng/ml) at various times after serum stimulation and similarly analyzed. Cells from each culture were collected 20 hr after serum stimulation and their cell cycle profiles were analyzed by FACS. The percentage of S phase cells measured was normalized to the value of a serum stimulated and an otherwise untreated cell sample also collected 20 hr after serum stimulation.
DISCUSSION
E2F family members and RB play important roles in cell cycle regulation (reviewed in ref. 1). Certain E2F species, operating as transcription factors, normally drive cell proliferation, which is in keeping with the fact that overproduction of certain E2F species stimulates G1/S progression.
E2F-1, when overproduced, can force certain G0-arrested cells to enter the cell cycle (37, 38). Synthesis of E2F-1 commences during the G0 to S phase parade and then falls after S phase entry (39). A number of genes encoding cell cycle progression proteins appear to be activated by E2F-1, such as DNA Pol α, cyclin E, cdk 2, cdc6 (for references see ref. 40), and certain members of the Mcm-family (10), and these varied observations all suggest that timely E2F-1 synthesis during G0 exit is an important event in the exit process. The analysis of cells that can and cannot synthesize E2F-1, presented here, now shows that E2F-1 function is essential for timely S phase entry after G0 exit. Indeed, the kinetics of cyclin E synthesis paralleled those of G0 to S progression, underscoring the fact that timely E2F-1 synthesis during G0 exit contributes in a significant way to cell cycle progression (41).
These results indicate that there is a unique role for E2F-1 function during G0 exit, which is not substituted in MEFs by any other member of the E2F family. Cyclin E RNA synthesis is an E2F-dependent event, and the kinetics of cyclin E synthesis are a dominant factor in the determining the kinetics of G0 exit (8, 29, 30). Hence, it seems fair to hypothesize that E2F-1 synthesis is a rate limiting event in the normal activation of the cyclin E promoter during the G0 exit process.
Consistent with what has been published previously (27, 42), no change in cell cycle intervals was observed among cycling MEF populations that do and do not synthesize E2F-1. Hence, in keeping with prior results, one might hypothesize that there are fundamental differences that govern the mechanisms of G1 exit in cells, which began this process in G0 as opposed to those that did so at the M/G1 boundary and never exited from the cell cycle.
Another feature of the nullizygous cells was that they displayed normal restriction point control. Thus, E2F-1 does not contribute, in a unique way, to the maintenance of restriction point regulation. By contrast, it plays an individuated role in events after the restriction point, given that it took longer to reach S phase from the restriction point in nullizygous than in wild-type cells. This conclusion fits with a model in which free E2F-1 function translates into the promotion of G1 progression after the restriction point, whereas phosphorylation of RB, an event that results in the release of free E2F-1/DP complexes, is part of the mechanism that determines whether and when restriction point regulation occurs (33).
The cell cycle entry promotion function of E2F-1 does not explain its role as a tumor suppressing agent in mice. In one model that could explain the latter function, it has been argued that E2F-1 acts as a component of an RB/E2F complex dedicated, in part, to normal proliferation control (42–44). In another, based again on evidence obtained in E2F-1 knockout mice, it can be shown that loss of E2F-1 function is linked to a defect in apoptosis of certain cell populations (27, 43, 44). Furthermore, we have recently observed that E2F-1, when overproduced, can have a negative effect on cell proliferation and cell cycle progression (Z.M.W., H.Y., F. Martelli, and D.M.L., unpublished work). Whatever the detailed mechanism of the tumor suppression effect, there is a dichotomy between the function of E2F-1 as a cell cycle entry promotion agent and a relatively long latency tumor suppressor.
Acknowledgments
We thank our laboratory colleagues for numerous helpful discussions, M. A. Thompson and M. Greenberg for providing E2F-1 embryos, and members of the Dana–Farber Cancer Institute flow cytometry facility for their expert help. Z.M.W. was a recipient of a postdoctoral fellowship from the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation (DRG-1334). H.Y. was supported by a generous grant from the Sharf-Green family fund. This work was supported by grants from the National Cancer Institute and the Dana–Farber/Novartis Drug Development Program.
ABBREVIATIONS
- MEF
mouse embryo fibroblast
- FACS
fluorescence-activated cell sorter
- RB
retinoblastoma
References
- 1.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 2.Cobrinik D. Curr Top Microbiol Immunol. 1996;208:31–61. doi: 10.1007/978-3-642-79910-5_2. [DOI] [PubMed] [Google Scholar]
- 3.La Thangue N B. Biochem Soc Trans. 1996;24:54–59. doi: 10.1042/bst0240054. [DOI] [PubMed] [Google Scholar]
- 4.Lam E W, La Thangue N B. Curr Opin Cell Biol. 1994;6:859–866. doi: 10.1016/0955-0674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 5.Nevins J R, Leone G, DeGregori J, Jakoi L. J Cell Physiol. 1997;173:233–236. doi: 10.1002/(SICI)1097-4652(199711)173:2<233::AID-JCP27>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 6.Slansky J E, Farnham P J. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 7.DeGregori J, Kowalik T, Nevins J R. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng Y, Eaton E N, Picon M, Roberts J M, Lundberg A S, Gifford A, Sardet C, Weinberg R A. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- 9.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamel P A, Gill R M, Phillips R A, Gallie B L. Mol Cell Biol. 1992;12:3431–3438. doi: 10.1128/mcb.12.8.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weintraub S J, Prater C A, Dean D C. Nature (London) 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 13.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Nature (London) 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 14.Sellers W R, Rodgers J W, Kaelin W J. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bremner R, Cohen B L, Sopta M, Hamel P A, Ingles C J, Gallie B L, Phillips R A. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Means A L, Slansky J E, McMahon S L, Knuth M W, Farnham P J. Mol Cell Biol. 1992;12:1054–1063. doi: 10.1128/mcb.12.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wade M, Kowalik T F, Mudryj M, Huang E S, Azizkhan J C. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taya Y. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang J Y. Curr Opin Genet Dev. 1997;7:39–45. doi: 10.1016/s0959-437x(97)80107-4. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann F, Martelli F, Livingston D M, Wang Z Y. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 21.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 22.Campanero M R, Flemington E K. Proc Natl Acad Sci USA. 1997;94:2221–2226. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krek W, Ewen M E, Shirodkar S, Arany Z, Kaelin W J, Livingston D M. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 24.Krek W, Xu G, Livingston D M. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 25.Dynlacht B D, Flores O, Lees J A, Harlow E. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 26.Xu M, Sheppard K A, Peng C Y, Yee A S, Piwnica W H. Mol Cell Biol. 1994;14:8420–8431. doi: 10.1128/mcb.14.12.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Field S J, Tsai F Y, Kuo F, Zubiaga A M, Kaelin W, Jr, Livingston D M, Orkin S H, Greenberg M E. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 28.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 29.Ohtsubo M, Roberts J M. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- 30.Dulic V, Lees E, Reed S I. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 31.Pardee A B. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 32.Bartek J, Bartkova J, Lukas J. Curr Opin Cell Biol. 1996;8:805–814. doi: 10.1016/s0955-0674(96)80081-0. [DOI] [PubMed] [Google Scholar]
- 33.Herrera R E, Sah V P, Williams B O, Makela T P, Weinberg R A, Jacks T. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herrera R E, Makela T P, Weinberg R A. Mol Biol Cell. 1996;7:1335–1342. doi: 10.1091/mbc.7.9.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zetterberg A, Larsson O, Wiman K G. Curr Opin Cell Biol. 1995;7:835–842. doi: 10.1016/0955-0674(95)80067-0. [DOI] [PubMed] [Google Scholar]
- 36.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 37.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Nature (London) 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 38.Qin X Q, Livingston D M, Kaelin W G, Jr, Adams P D. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandara L R, Buck V M, Zamanian M, Johnston L H, La Thangue N B. EMBO J. 1993;12:4317–4324. doi: 10.1002/j.1460-2075.1993.tb06116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nevins J R. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 41.Ohtani K, DeGregori J, Nevins J R. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 43.Tsai K, Hu Y, Macleod K, Crowley D, Yamasaki L, Jacks T. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 44.Pan H, Yin C, Dyson N J, Harlow E, Yamasaki L, Van Dyke T. Mol Cell. 1998;2:283–292. doi: 10.1016/s1097-2765(00)80273-7. [DOI] [PubMed] [Google Scholar]