Abstract
A visible light photoinitiator, eosin, in combination with a tertiary amine coinitiator is found to initiate polymerization despite the presence of at least 1000-fold excess dissolved oxygen which functions as an inhibitor of radical polymerizations. Additionally, 0.4 µM eosin is able to overcome 100-fold excess (40 µM) 2,2,6,6-Tetramethyl-1-piperidinyloxy (TEMPO) inhibitor, initiating polymerization after only a 2 minute inhibition period. In contrast, 40 µM Irgacure-2959, a standard cleavage-type initiator, is unable to overcome even an equivalent amount of inhibitor (40 µM TEMPO). Through additional comparisons of these two initiation systems, a reaction mechanism is developed which is consistent with the kinetic data and provides an explanation for eosin’s relative insensitivity to oxygen, TEMPO and other inhibitors. A cyclic mechanism is proposed in which semi-reduced eosin radicals react by disproportionation with radical inhibitors and radical intermediates in the inhibition process to regenerate eosin and effectively consume inhibitor. In behavior similar to that of eosin, rose bengal, fluorescein, and riboflavin are also found to initiate polymerization despite the presence of excess TEMPO, indicating that cyclic regeneration likely enhances the photoinitiation kinetics of many dye photosensitizers. Selection of such dye initiation systems constitutes a valuable strategy for alleviating inhibitory effects in radical polymerizations.
Keywords: photopolymerization, initiators, hydrogels, inhibition, eosin
Introduction
The ability to overcome inhibition is critical to nearly all applications that rely on radical polymerization. In particular, radical-mediated photopolymerization reactions are ubiquitous and highly capable,1 yet they suffer from significant inhibitory effects that limit their implementation or make that implementation more costly. Common inhibitors include impurities present in components of the monomer formulation, intentionally added inhibitors such as hydroquinones that are used to stabilize the monomer by preventing unintended polymerization, and dissolved oxygen, which is introduced to the monomer by the atmosphere.2,3 Polymerization is unable to proceed until the inhibitor concentration has been reduced below a critical level whereupon initiation and propagation become more facile than the inhibitory reactions, at which point the radicals preferentially react with monomer. During a polymerization reaction, inhibitor is readily consumed by reactions with initiating and propagating radicals. If sufficient radicals are generated, and if the inhibitor is nonreplenishable, then polymerization will proceed after an inhibition period, though often at a reduced rate. However, if an insufficient number of initiating radicals are generated, polymerization will never occur as is generally the case when the inhibitor concentration is greater than the initiator concentration. Especially problematic is polymerization in ambient atmosphere as oxygen continually diffuses back into the resin, resulting in a tacky surface due to incomplete cure associated with inhibition.
One of the most common methods for dealing with inhibitors is to use initiators at concentrations well beyond that of the inhibitors; however, as initiator concentration is increased, difficulties arise with light attenuation, yellowing of the resin, instability of the resulting polymer, reduction of the material properties and cost increases, all of which limit the feasibility of high initiator concentrations.1,4 When the initiator is in excess of the inhibitor, the inhibition time is reduced by employing higher light intensities which yield more rapid radical generation, but this outcome is achieved at the cost of increased energy requirements, additional processing demands, and potential changes to the final polymer properties. In an alternate approach, inhibitor concentrations may be reduced prior to polymerization by additionally purifying the monomer, though this approach requires further processing steps and complete removal of inhibitor is impractical. The dissolved oxygen concentration is often reduced by interaction with an inert purge gas in one of many forms.5,6 Unfortunately, use of an inert purge gas is both costly and adds complexity to the process. An additional approach to address inhibition is to employ monomer additives or monomer formulations with unique kinetic capabilities. For example, tertiary amines and vinyl pyrrolidone (VP) are monomer additives that have been shown to mitigate the effects of oxygen inhibition.7–9 Thiol-ene and thiol-acrylate monomer formulations, which undergo a radically mediated step growth process, are relatively insensitive to oxygen inhibition, yet due to the reduced modulus of these materials, there are limitations to the applications in which they are suitable.10–12
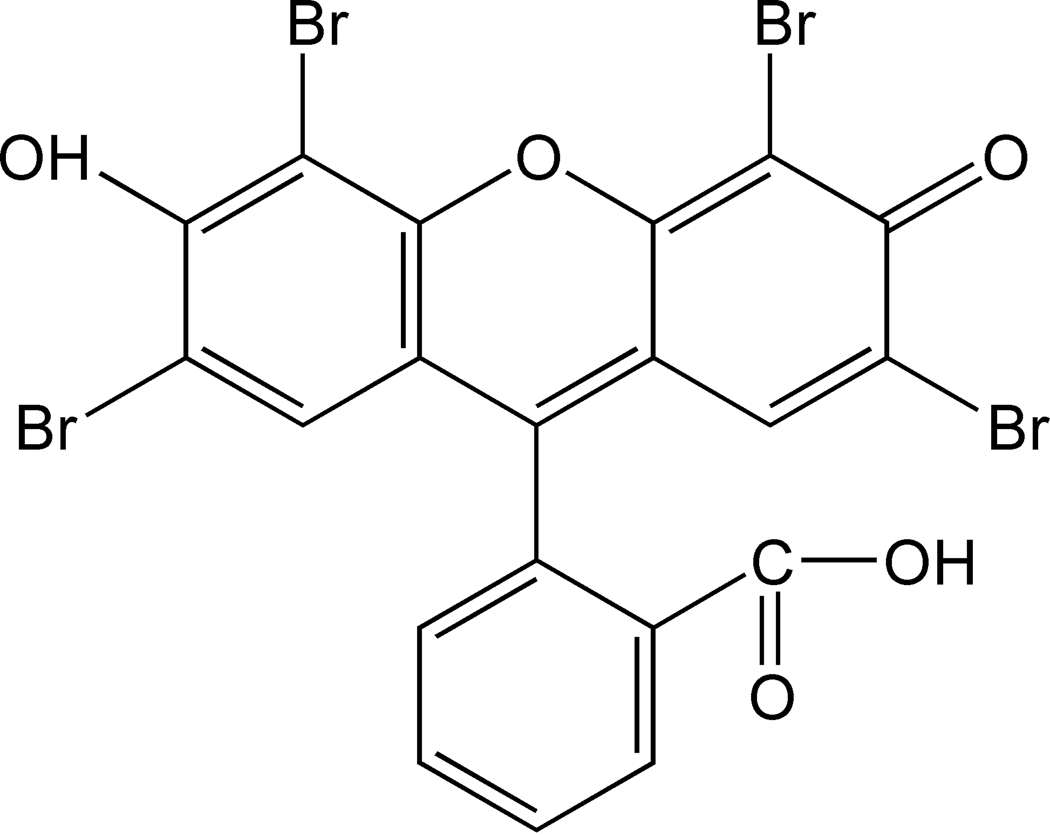
Though a variety of methods have been developed to address the problem of polymerization inhibition, identification of additional, novel techniques for overcoming general inhibitory effects would be beneficial, especially if the new approaches were simple and cost effective to implement. Here, we present evidence for and a proposed mechanism by which eosin (Figure 1), a visible light photoinitiator, in combination with a tertiary amine coinitiator is able to initiate polymerization in the presence of an extreme excess of inhibitor, suggesting that use of dye photosensitizer initiation systems may inherently help reduce inhibitory effects.
Figure 1.
Eosin chemical structure.
Photoinitiators that absorb in the visible as opposed to the UV range are advantageous in multiple applications because visible light enhances cure depth, is less damaging to biological samples, and is safer for technicians and users who are at risk of exposure to the light source. Many visible light-initiated photopolymerizations are achieved with type II photoinitiation systems which typically comprise a dye photosensitizer and a coinitiator, commonly a tertiary amine. The dye photosensitizer absorbs light, then undergoes energy, charge, and/or electron transfer with the coinitiator, yielding a semi-reduced radical dye species and a coinitiator radical capable of initiating polymerization.9 Eosin is commercially available and has been employed for a wide variety of applications including cell encapsulation,13 creation of hydrogel biomaterials for in vivo use with tissues,14 surface modification of nanoparticles,15 manufacturing of holographic data storage material,16 and biodetection via polymerization-based signal amplification.17,18 A variety of other work has been performed with eosin and eosin-derivatives for initiating polymerizations that focuses on improving or altering various aspects of the initiation reaction.19–22
In this communication we present observations that eosin with a tertiary amine coinitiator is able to overcome at least 1000-fold excess oxygen and 100-fold excess 2,2,6,6-Tetramethyl-1-piperidinyloxy (TEMPO) inhibitor. To validate and ascertain the explanation for these observations, similar experiments were performed with a cleavage-type initiator, Irgacure 2959 (I2959), which as expected, was prevented from initiating polymerization by the presence of excess inhibitor. Through kinetic analysis of eosin- and I2959-initiated polymerizations, a reaction mechanism is presented which takes into account the kinetic data and provides an explanation for eosin’s ability to overcome oxygen and TEMPO inhibitors. The proposed mechanism posits that semi-reduced eosin radicals react with radical inhibitors or radical intermediates in the inhibition process to regenerate eosin and consume inhibitor, thereby enabling eosin to overcome an extreme excess of inhibitors through a cyclic reaction process. To assess whether other dye photosensitizers share this reaction behavior, rose bengal, fluorescein, riboflavin, camphorquinone and benzophenone are investigated for their ability to initiate polymerization in the presence of TEMPO.
Although a handful of regenerative reactions for dye molecules have been presented previously,9 to our knowledge, no previous reports have demonstrated the ability of a dye/amine photoinitiation system to overcome a large excess of polymerization inhibitors. A few publications have discussed the use of eosin with ascorbic acid as a coinitiator to polymerize acryamide,23,24 with the observation that oxygen is actually required for polymerization to proceed, though no information was provided concerning other types of inhibitors. In contrast, the eosin/tertiary amine results presented here indicate a clearly distinct mechanism, as evidenced by the fact that oxygen is exclusively inhibitory. Further, as tertiary amines are a more commonly used coinitiator than ascorbic acid, the mechanism posited here may be more relevant to a wider variety of applications.
Experimental
Materials
Poly(ethylene glycol) diacrylate (PEGDA) (Mn = 575), N-methyldiethanolamine (MDEA), 1-vinyl-2-pyrrolidone (VP), 2,2,6,6-Tetramethyl-1-piperidinyloxy (TEMPO), rose bengal, fluorescein, benzophenone, camphorquinone, riboflavin, streptavidin, and sodium carbonate were purchased from Sigma-Aldrich. PEGDA was purified twice using columns from Scientific Polymer Products that remove hydroquinone monomethyl ether (MEHQ). Eosin-isothiocyanate (eosin-ITC) was purchased from Invitrogen. Irgacure 2959 (I2959) (2-Hydroxy-1-[4-(2-hydroxyethoxy) phenyl]-2-methyl-1-propanone) was obtained from Ciba Specialty Chemicals. Eosin-conjugated streptavidin (SA-eosin) was synthesized as described previously.25 Silicone gasket material was purchased from Grace Bio-Labs, and water was purified using a Milli-Q system.
Since one of the potential applications for eosin is polymerization-based-amplification where overcoming trace inhibitors is critical to the development of an ultrasensitive detection platform,24,26 we have evaluated eosin both in its non-conjugated, eosin-isothiocyanate (eosin-ITC) form as well as in an eosin-streptavidin conjugate (SA-eosin), prepared as previously described.25 Previously published results report that in an aqueous PEGDA formulation, initiation with SA-eosin as compared to eosin-ITC resulted in only a slightly decreased polymerization rate.24 It is not anticipated that the presence of streptavidin or the isothiocyanate group has a significant effect on the initiation behavior of eosin under the conditions investigated here.
Polymerizations
A Nicolet Magna-IR 760 E.S.P. infrared spectrometer adapted with a horizontal sample holder was used to monitor polymerizations in real time, tracking the peak centered at approximately 6175 cm−1. C=C conversion was calculated by the change in peak area. Sample chambers were prepared using binder clips to clamp two glass slides on either side of a 0.5 mm silicone gasket cut to have a 2 cm × 1 cm hole for monomer. Unless otherwise indicated, argon was bubbled through monomer for 20 minutes, and then monomer was introduced to the sample chamber with a syringe and needle. All experiments were performed with PEGDA (420 mM) and VP (35 mM) in water. For eosin-initiated polymerizations, MDEA (210 mM) was also included as a coinitiator. Visible light irradiation of eosin and rose bengal was achieved with an Acticure (Exfo) high pressure mercury lamp with an in-house internal bandpass filter (350 nm – 650 nm) and an external 490 nm longpass filter (Edmund Optics) positioned at the end of a light guide and a collimating lens. A similar light setup was used for UV irradiation of I2959 and benzophenone, except that no external filter was used and the internal filter was a 320–390 nm bandpass filter (Exfo). For excitation of camphorquinone, fluorescein and riboflavin, a 400–500 nm bandpass filter (Exfo) was used. Light intensities were measured by an International Light radiometer. Each condition was tested in triplicate.
Results and Discussion
Resistance of Eosin to Oxygen Inhibition
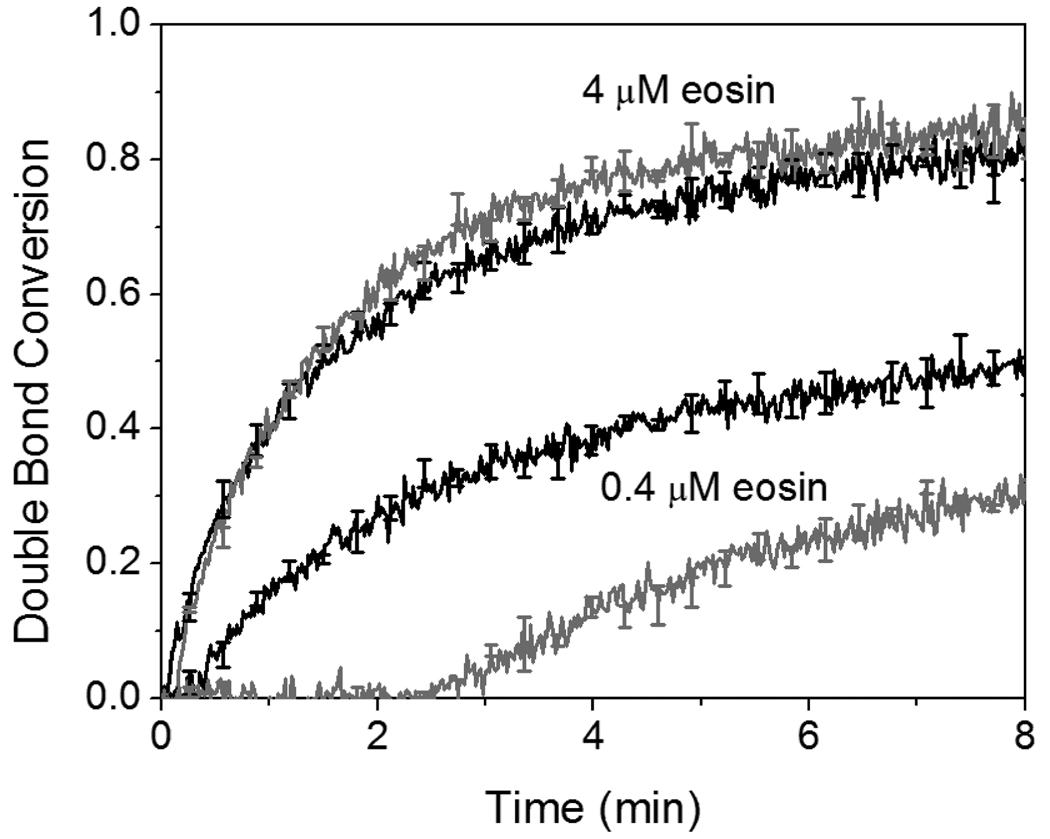
Oxygen inhibition of polymerization is a common problem in radical polymerization, resulting in longer inhibition times and slower polymerization rates. Typically, for samples in contact with air, dissolved oxygen concentrations in acrylic monomers are found to be in the 0.6 – 2 mM range27 while the dissolved oxygen concentration in water is expected to be approximately 0.3 mM.28 Despite the relatively high dissolved oxygen concentrations in an aqueous 420 mM PEGDA formulation containing 210 mM MDEA as a coinitiator and 35 mM VP, as little as 0.4 µM eosin is able to initiate polymerization in the presence of dissolved oxygen after a less than 3 minute inhibition period, demonstrating that eosin is capable of overcoming on the order of at least 1000-fold excess inhibitor (Figure 2). Additionally, as shown in Table 1, polymerization by 4 µM eosin, showed no change in polymerization rate and a less than 2-fold increase in inhibition time (tinh) when no measures were taken to reduce the dissolved oxygen content in equilibrium with ambient air. Similar experiments were performed with I2959, a type I α-hydroxy ketone photoinitiator, in an aqueous 420 mM PEGDA formulation containing 35 mM VP and no MDEA. Here, it was found that even 4 mM I2959 (104-fold higher than the eosin concentration!) displays an almost 6-fold increase in tinh and a 2-fold decrease in polymerization rate when monomer is not purged with argon as compared to when purging is performed. Although MDEA is a common additive for reducing the effect of oxygen inhibition,9 it was found that MDEA alone is not responsible for the drastically reduced sensitivity to oxygen seen with the eosin system, as evidenced by the marked inhibition still present with 0.4 mM I2959 when 210 mM MDEA was added to the monomer formulation (Table 1). In summary, these data indicate that eosin has a strong resistance to oxygen inhibition, particularly at extremely low eosin concentrations.
Figure 2.
Comparison of the reaction kinetics of eosin-initiated polymerization with monomer that has been purged with argon for 20 minutes immediately prior to reaction (black line) versus monomer that has not been treated to reduce dissolved oxygen content (grey line). The initiator is eosin-ITC and the monomer is 420 mM PEGDA, 210 mM MDEA, and 35 mM VP in water. Samples were in a sealed 0.5 mm thick sample holder and were exposed to 75 mW/cm2 >480 nm light and the light was turned on at time equal to zero in all experiments.
Table 1.
aEffect of dissolved oxygen on Rp and tinh in polymerization initiated by eosin or I2959.
| Concentration | Rp | tinh | tinh no argon | ||
|---|---|---|---|---|---|
| Initiator | (M) | Argon | (M/s) | (s) | tinh with argon |
| Eosin | 4 × 10−6 | yes | 7.2 (+/− 1.1) × 10−3 | 5.6 (+/− 0.4) | 1.8 |
| + MDEA | no | 6.2 (+/− 0.6) × 10−3 | 10 (+/− 1) | ||
| Eosin | 4 × 10−7 | yes | 1.8 (+/− 0.2) × 10−3 | 31 (+/− 2) | 6.1 |
| + MDEA | no | 7.5 (+/− 0.5) × 10−4 | 190 (+/− 10) | ||
| I2959 | 4 × 10−3 | yes | 5.0 (+/− 0.3) × 10−2 | 9.0 (+/− 0.6) | 5.8 |
| no | 2.4 (+/− 0.2) × 10−2 | 52 (+/− 2) | |||
| I2959 | 4 × 10−4 | yes | 1.9 (+/− 0.2) × 10−2 | 10 (+/−3) | 8.7 |
| + MDEA | no | 1.3 (+/− 0.2) × 10−2 | 87 (+/−7) | ||
Polymerization rates were calculated for the region encompassing 5–30% conversion. Eosin-ITC-initiated reactions were performed with 420 mM PEGDA, 210 mM MDEA, and 35 mM VP in water exposed to 75 mW/cm2 light at wavelengths greater than 480 nm. I2959-initiated reactions were performed with the same monomer formulation, except 210 mM MDEA was only used as indicated. Irradiation conditions for I2959 initiation were 10 mW/cm2, 320–390 nm light. When indicated, monomer was bubbled with argon for 20 minutes immediately prior to polymerization.
Resistance of Eosin to TEMPO inhibition
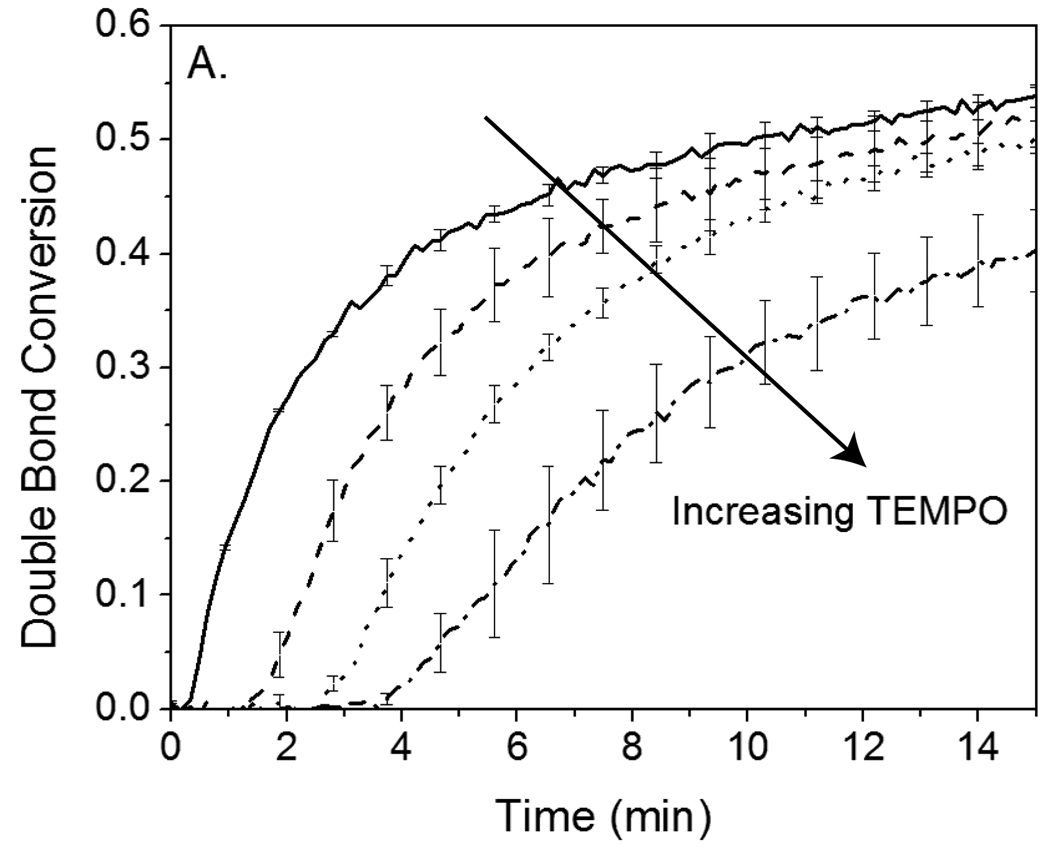
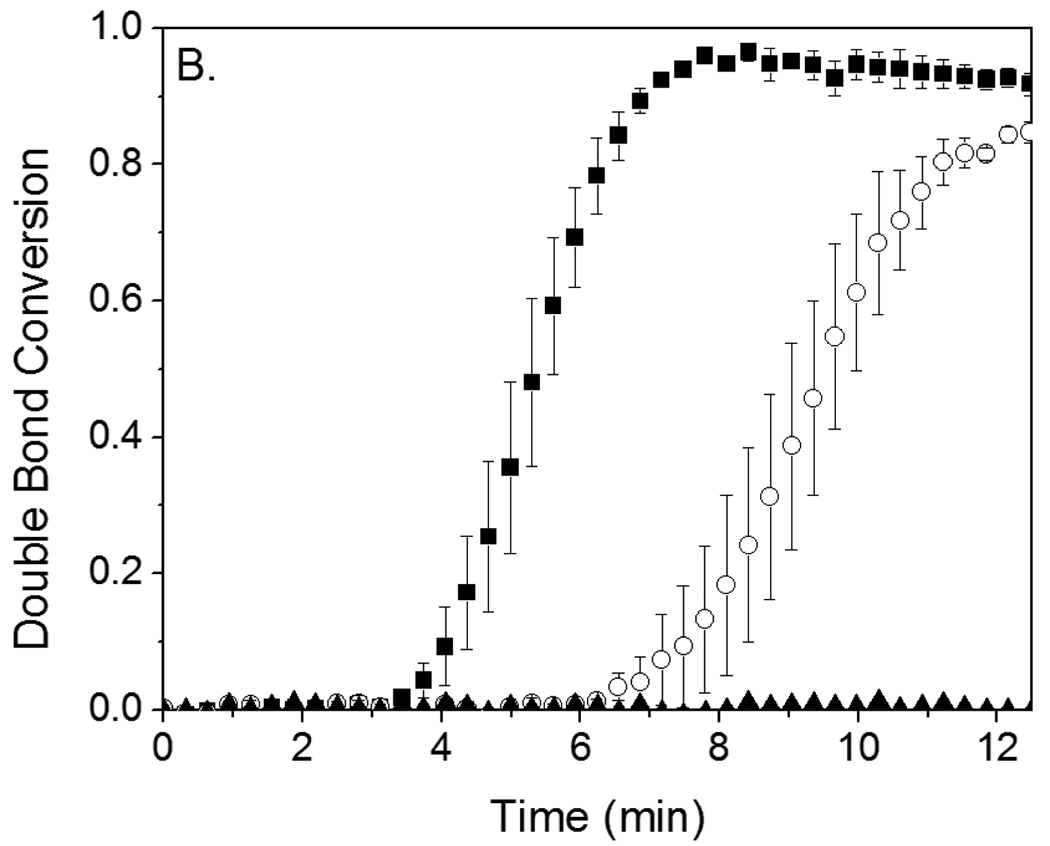
To ascertain whether eosin’s behavior was specific to overcoming oxygen inhibition or more general, eosin-initiated polymerizations were performed in the presence of TEMPO , thereby evaluating eosin’s ability to overcome a second inhibitor type. These experiments also enabled the eosin initiation rate (Ri) determination. TEMPO is a nitroxide radical known for its role in mediating controlled polymerizations at elevated temperatures (above 120°C), though at lower temperatures it functions predominantly as a polymerization inhibitor, reacting rapidly with any carbon-based radicals generated by the initiating system.2,29,30 Since both initiating and propagating radicals react with TEMPO much more preferentially than with monomer, polymerization does not proceed until TEMPO is essentially completely consumed. Thus, measurement of the time required for a known concentration of TEMPO to be consumed (tinh) enables determination of the radical generation rate, Ri. Consistent with the oxygen inhibition results, polymerizations initiated by eosin required on the order of 100-fold excess TEMPO compared to eosin before a significant increase in tinh was observed. As shown in Figure 3A, 0.4 µM eosin is able to initiate polymerization in the presence of 60 µM TEMPO after only a 4 minute inhibition period. In contrast, even after 12 minutes, 40 µM I2959 is not able to initiate polymerization in the presence of 40 µM TEMPO (Figure 3B), and with 4 µM TEMPO, polymerization is able to proceed only after an approximately 2 minute increase in tinh. To ascertain whether MDEA contributes to eosin’s ability to overcome high concentrations of TEMPO, 210 mM MDEA was added to a polymerization initiated by 40 µM I2959 in the presence and absence of 4 µM TEMPO. It was found that even with MDEA present, a 2 minute increase in inhibition time from 1.4 (+/− 0.3) min to 3.4 (+/− 0.7) min was still observed upon addition of TEMPO. The fact that eosin initiates polymerization in the presence of 1000-fold excess oxygen and 100-fold excess TEMPO indicates that each eosin molecule must be capable of generating a very large number of initiating radicals over time.
Figure 3.
A. Photopolymerization kinetics initiated by 0.4 µM eosin (as a component of SA-eosin) in the presence of 0, 20, 40, and 60 µM TEMPO inhibitor. B. Reaction kinetics of polymerization initiated by 40 µM I2959 in the presence of 0 (■), 4 (○), and 40 (▲, data points all fall along the x-axis at near-zero conversion) µM TEMPO inhibitor. Monomer consists of an aqueous solution of 420 mM PEGDA, 35 mM VP and, in the case of the eosin-initated systems, 210 mM MDEA as a coinitiator. Eosin was irradiated with 75 mW/cm2 >480 nm light, while I2959 was exposed to 10 mW/cm2, 320–390 nm light.
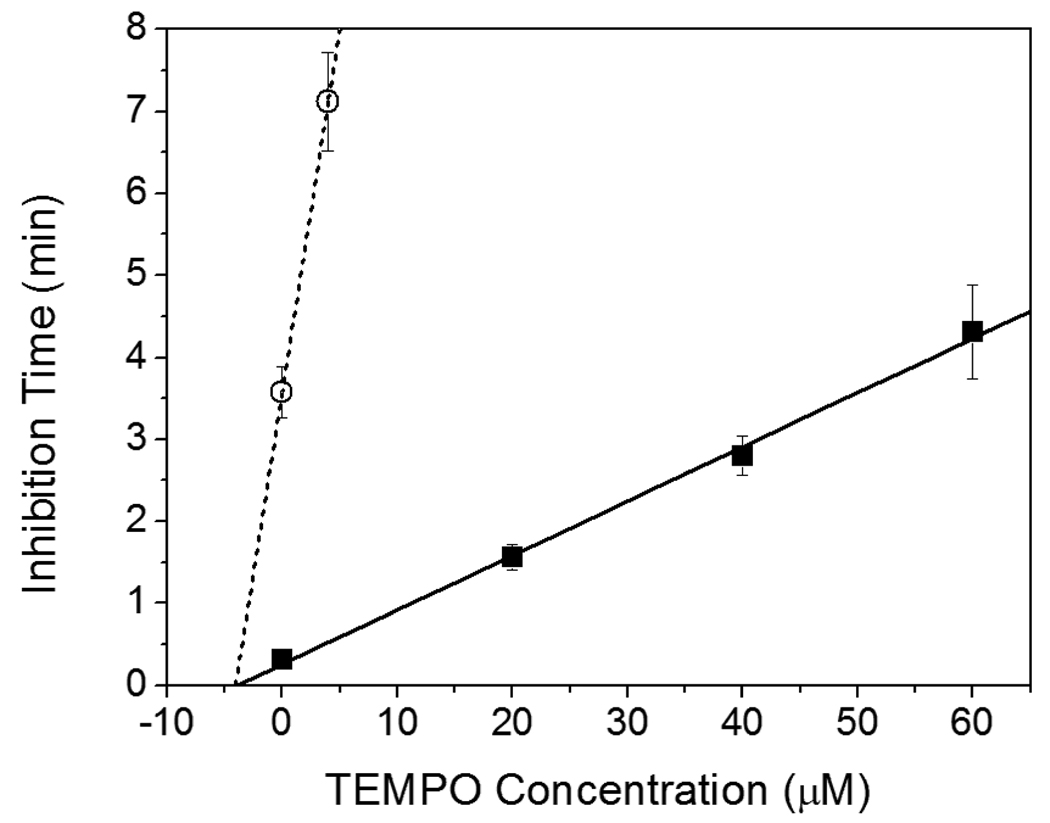
To determine Ri , the average initiation (or radical generation) rate during the inhibition period, for the eosin and I2959 initiated polymerizations depicted in Figure 3, the TEMPO concentration was plotted against tinh (Figure 4). Ri, calculated as the inverse slope of these lines ([TEMPO]/tinh), was found to be 2.3 (+/− 0.2) × 10−7 M/s for the eosin system and 1.6 (+/− 0.3) × 10−8 M/s for the I2959 system. It appears at first unexpected that 40 µM I2959 would yield a lower Ri than 0.4 µM eosin, since the latter has a 100-fold lower initiator concentration though the difference is associated with the different intensities and molar absorptivities of these systems. A noteworthy feature of the data plotted in Figure 4 is that both lines have very similar x-intercepts, calculated to be 3.8 (+/− 1.4) µM for the eosin experiments and 3.3 (+/− 1.1) µM for the I2959 experiments. As the x-intercept represents the inhibitor concentration present in the monomer prior to addition of TEMPO, it is expected that this value should be the same for both systems, since the same monomer was used in each case. Thus, the fact that both lines have the same x-intercept further verifies the validity of these results.
Figure 4.
Adaptation of the data in Figure 3 to show the correlation of inhibition time with TEMPO concentration in both the eosin system (■ and —) and the I2959 system (○ and ····). The inverse slopes of the lines indicate the initiation rate, or the radical generation rate, of each system. The x-intercept is indicative of the inhibitor concentration in the monomer prior to the addition of TEMPO.
Evidence for cyclic regeneration of eosin
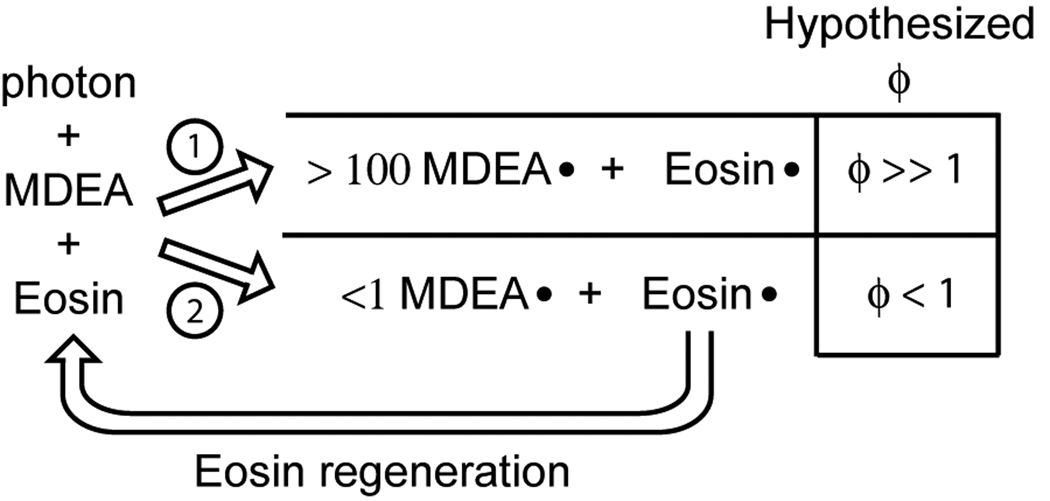
The observation that eosin is able to initiate polymerization in the presence of 100- to1000-fold excess inhibitor, signifies that each eosin molecule generates at least 100 to 1000 initiating radicals, prior to the polymerization starting and continues to generate sufficient radicals to promote rapid polymerization after the inhibition time is complete. Two scenarios may be posited in which an eosin molecule could generate many radicals (Figure 5): 1) upon absorbing a single photon, eosin would trigger a chain reaction in which many radicals would ultimately be generated; or, 2) upon absorbing a single photon eosin may generate an eosin radical and an MDEA radical, then eosin subsequently is regenerated, after which eosin may again absorb a photon and generate another MDEA radical, etc., such that at least one photon is absorbed for each MDEA radical created. To distinguish between these two scenarios, the rate of light absorption (Ia) and the initiation efficiency (ϕ= Ri/Ia) for the eosin and I2959 systems depicted in Figure 3 and Figure 4 were calculated and are shown in Table 2. Ia was calculated by the Beer-Lambert law [Ia = Io (1−10−εbC)] where Io is the incident light as measured by a radiometer, ε is the molar absorptivity of the photoinitiator, b is the thickness of the sample (0.5 mm) and C is the concentration of the photoinitiator. In the case of eosin Ia was 1.5 × 10−6 M/s, and ϕ was 0.16, whereas with I2959, Ia was 8.7 × 10−7 M/s and ϕ was 0.02. The low values for ϕ are likely due in part to the very low monomer concentration (420 mM PEGDA), which would make side reactions of the primary radicals relatively more prevalent than the primary radical reaction with monomer. The fact that ϕ for eosin is significantly less than 1 supports the hypothesis that eosin generates multiple radicals through a process in which eosin is cyclically regenerated.
Figure 5.
Two scenarios by which eosin would be able to generate multiple initiating radicals. The corresponding hypothesized initiation efficiency (ϕ = Ri/Ia) is indicated for both scenarios.
Table 2.
Comparison of various kinetic parameters for eosin-initiated and I2959-initiated systems as calculated from the experiments depicted in Figure 2 and Figure 3.
| Ri (M/s) | Ia (M/s) | ϕ (Ri/Ia) | Rp (M/s) | |
|---|---|---|---|---|
| Eosin | 2.3 (+/− 0.2) × 10−7 | 1.5 × 10−6 | 0.16 | 1.8 (+/− 0.1) ×10−3 |
| I2959 | 1.6 (+/− 0.3) × 10−8 | 8.7 × 10−7 | 0.02 | 4.1 (+/− 0.4) × 10−3 |
Proposed mechanism for cyclic eosin regeneration
Further analysis of the data presented in Table 2 reveals that eosin initiation causes a higher termination rate as compared to I2959 initiation. Classically, assuming chain-length independent bimolecular termination, the polymerization rate (Rp) is dependent on Ri, monomer concentration [M], and the kinetic constants for termination (kt) and propagation (kp) according to: Rp = kp[M](Ri/2kt)0.5.2 [M], kp, and kt typically depend on the monomer formulation and are not generally expected to change with different radical initiators. Consequently, it is expected that Rp/Ri 0.5 will be equal for both the eosin- and the I2959-initiated systems. However, based on the Ri and Rp values measured in these experiments, this ratio is found to be over 8-fold higher in the-I2959 initiated system as compared to the eosin-initiated system. This indicates that Rp in the eosin system is not as high as would be expected for the measured Ri. The most likely explanation for this phenomenon is that termination is occurring more rapidly in the eosin-initiated system.
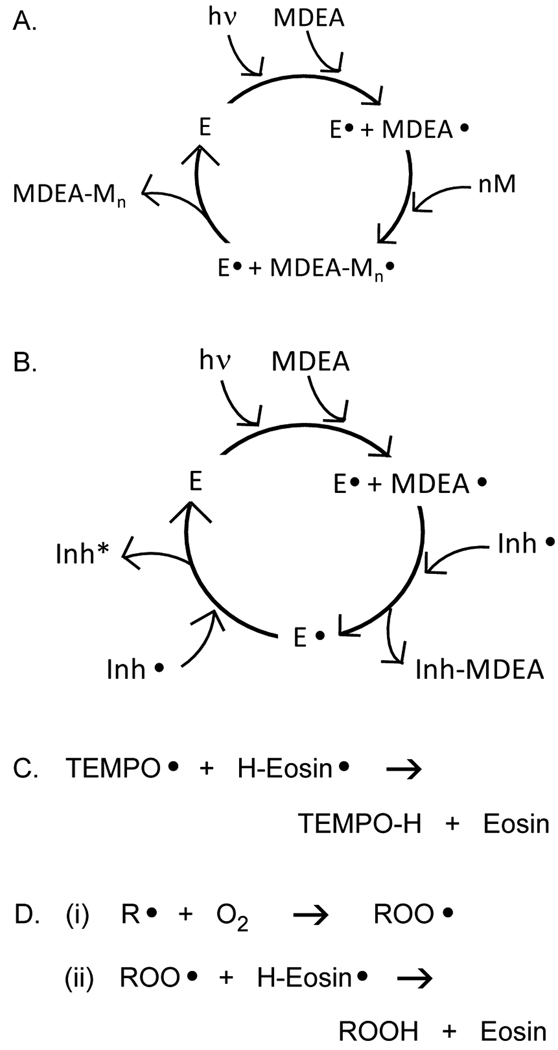
A possible cause of increased termination in the eosin system is primary radical termination. When eosin absorbs light, the triplet excited state of eosin undergoes a photoreductive hydrogen transfer with MDEA yielding a deprotonated initiating MDEA radical and a protonated eosin radical that is believed to be inactive for initiating, but active for termination.31,32 If the semi-reduced eosin radical were to undergo primary radical termination by disproportionation, then eosin would be regenerated, as depicted in the proposed mechanism in Figure 6A. This reaction, because the more mobile eosin radical would be expected to have a higher termination kinetic constant than a typical polymeric radical, would result in a slower polymerization than expected for the measured Ri.
Figure 6.
A. Proposed mechanism to explain the observation that eosin-initated polymerizations have a higher termination rate than expected. It is hypothesized that eosin radicals terminate propogating polymer chains by disproportionation, regenerating eosin and ultimately resulting in increased termination. B. Proposed mechanism by which eosin consumes inhibitors. It is hypothesized that eosin radicals react with inhibitor radicals or radical intermediates in the inhibition process, thereby regenerating eosin and consuming the inhibitor. Inactive inhibitor is depicted by Inh*. C and D. Specific proposed regeneration reactions of semi-reduced eosin with TEMPO and oxygen.
Likewise, if an eosin radical were to react by disproportionation with a stable radical inhibitor such as TEMPO, then eosin would be regenerated while the inhibitor would be consumed (Figures 6B and 6C), resulting in decreased sensitivity to the inhibitor and an ability to overcome inhibitor concentrations that were much higher than the initiator concentration. This posited mechanism is also applicable to eosin initiation in the presence of oxygen. When oxygen reacts with an initiating or propagating radical, it forms a peroxy radical (Figure 6Di) which reacts with monomer much more slowly than a propagating radical reacts with monomer, thereby effecting inhibition of polymerization.2 However, if an eosin radical were to react by disproportionation with the peroxy radical (Figure 6Dii), then eosin would be regenerated while the oxygen would still be a constituent of the resulting hydrogen peroxide and therefore inactive for additional inhibition reactions. The hypothesis that eosin radicals react with peroxy radicals to regenerate eosin is similar to a mechanism that has been put forth previously to explain the reduced oxygen sensitivity in thiol-ene and thiol-acrylate polymerizations. In the case of thiol-ene and thiol-acrylate polymerizations, peroxy radicals on the ends of terminated kinetic chains are believed to abstract hydrogen from thiols, yielding thiyl radicals that initiate new kinetic chains.10,11,33
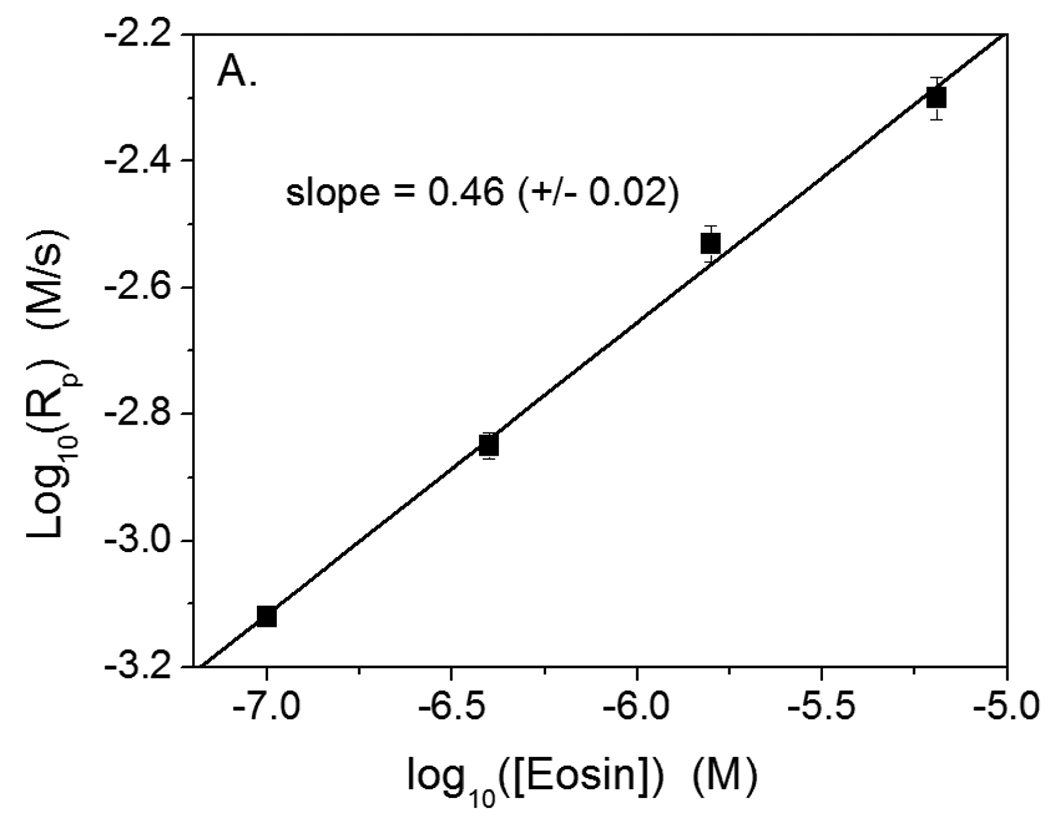
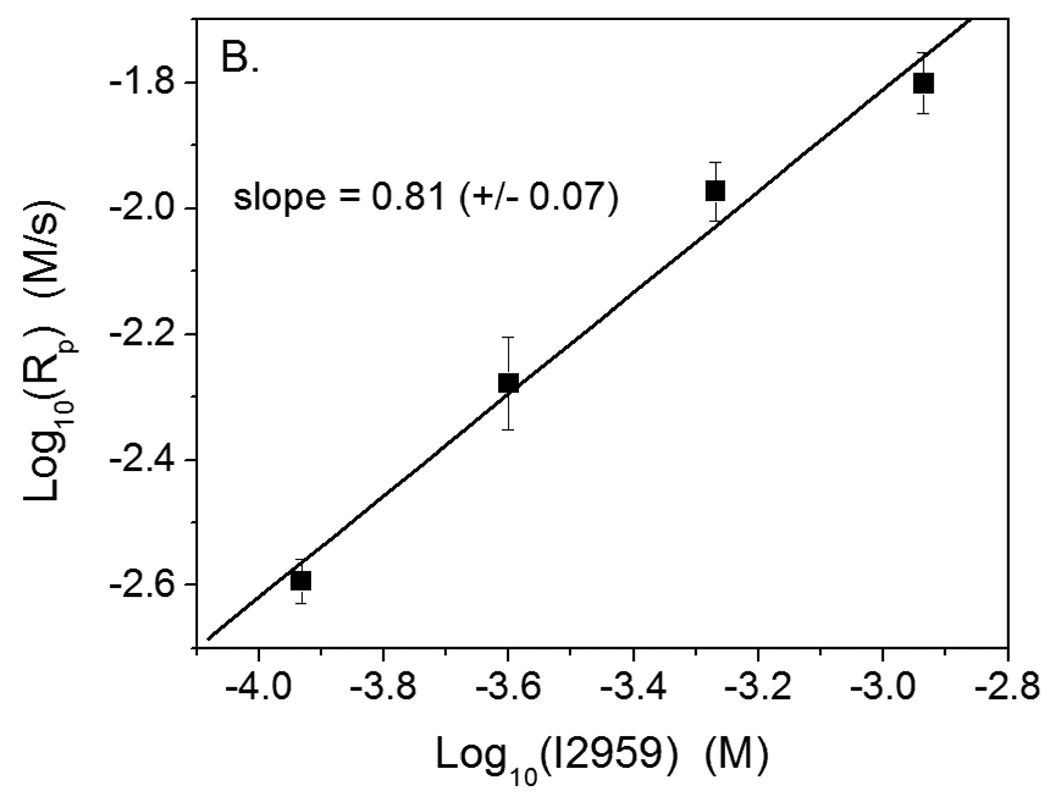
Further analysis and comparison of the polymerization rate in the eosin- and I2959-initiated polymerizations provides evidence in support of the hypothesis that eosin undergoes primary radical termination with propagating chains.. When chain length independent termination occurs predominately by bimolecular reaction of two propagating chains, then Rp ~ Ri α, with α equal to 0.5.2 However, when chain length dependent termination (i.e., short chains terminate more rapidly than long chains) occurs to a significant degree, α decreases, reaching a minimum of 0.3 in the particular case where one of the two primary radicals generated is completely inactive for initiation, but reactive towards termination.2,34 Here, α for the eosin system was found to be 0.46 (+/− 0.02) (Figure 7A), which is nearly the square root dependence that is expected classically in the absence of primary termination. However, for the same monomer system initiated by I2959, α was found to be 0.81 (+/− 0.07) (Figure 7B). MDEA was included in both the eosin- and I2959- initiated polymerizations to verify that any observed difference in α between these systems was not due to the presence of MDEA, which also acts as a chain transfer agent. The large α value in the I2959-initiated polymerizations indicates that the termination mode in the conventionally initiated system is dominated by radical trapping. This unimolecular termination reaction results from extremely low radical mobilities and causes the scaling exponent, α, to tend toward 1.0 instead of the traditional 0.5.4 The significant reduction in α when transitioning from the cleavage initiator to the eosin system is indicative of dramatic changes in the termination mechanism, as expected if the eosin radical is responsible for terminating most of the growing polymer chains by disproportionation as hypothesized. The value of 0.46 for α for the eosin system likely results from the interplay between both unimolecular termination and primary radical termination, the former tending to increase α and the latter tending to decrease it.
Figure 7.
Plots to calculate the scaling exponent between initiation rate and polymerization rate for polymerization in 420 mM PEGDA, 210 mM MDEA and 35 mM VP in water. A. Polymerization initiated by eosin-ITC with 75 mW/cm2 >480 nm light. B. Polymerization initiated by I2959 with 4.0 mW/cm2, 320–390 nm light. The lines represent linear, least squares fits to the experimental data. The slope of each line is α, which represents the scaling exponent between the polymerization and initiation rates.
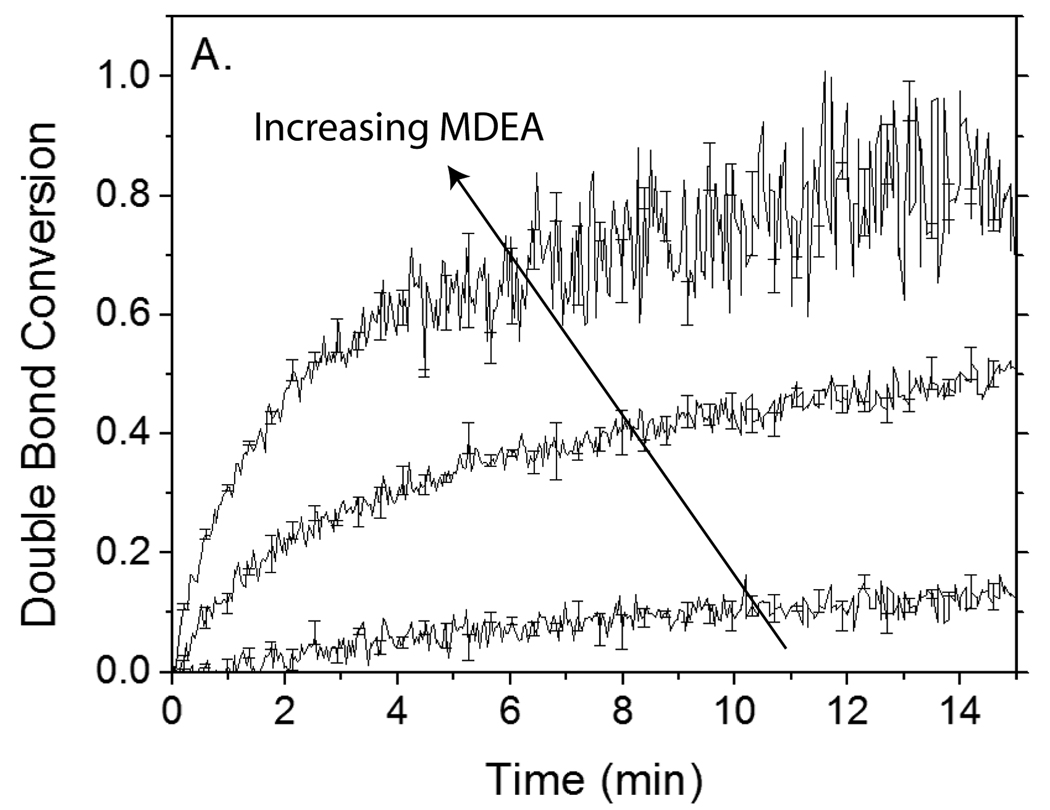
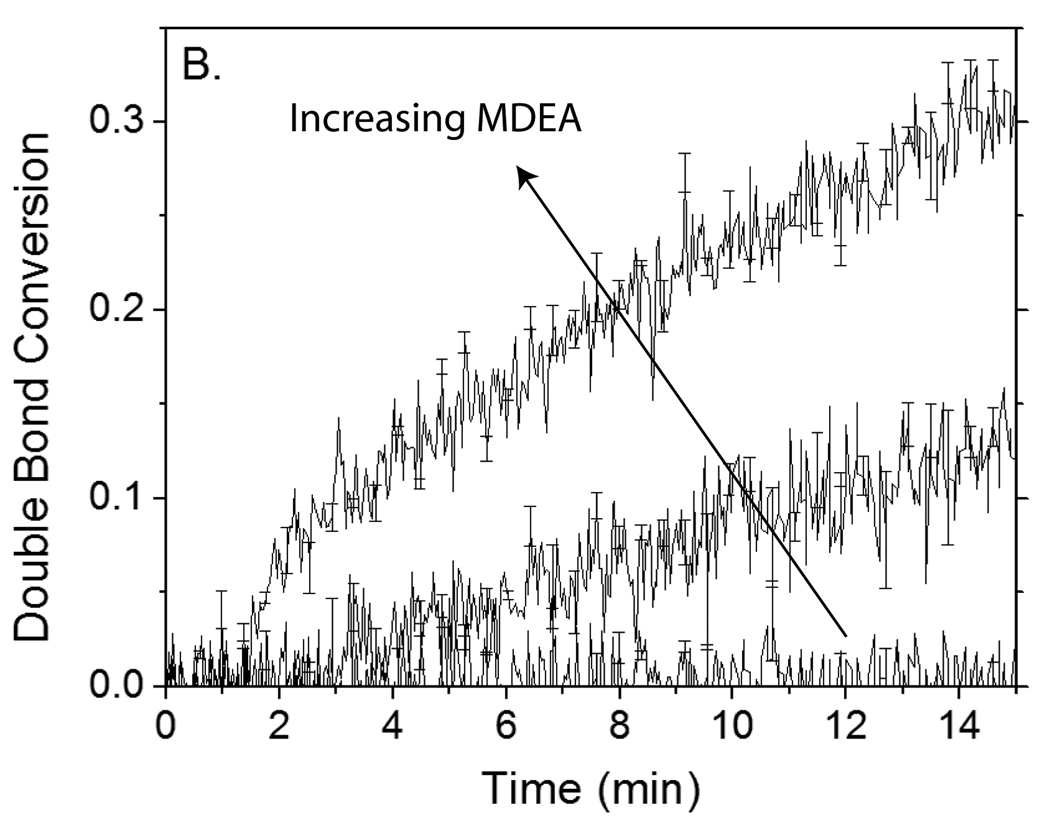
To evaluate the proposed eosin regeneration mechanism further, eosin’s initiation capability was investigated in the presence of 100-fold excess TEMPO at various MDEA concentrations. Since the mechanism posits that one MDEA molecule is consumed for each molecule of inhibitor that is inactivated, it is expected that eosin is incapable of initiating polymerization unless the MDEA concentration exceeds the TEMPO concentration. Additionally, as the MDEA concentration is increased, improved mitigation of TEMPO inhibition is anticipated. Consistent with these expectations, it is found that 4 µM eosin and 0.4 mM MDEA are unable to initiate polymerization in the presence of 0.4 mM TEMPO during the time frame investigated (Figure 8). However, increasing the MDEA concentration to 20-fold (8 mM) or 100-fold (40 mM) greater than the TEMPO concentration enables polymerization to occur readily after only a 3.3 and 1.5 min tinh, respectively.
Figure 8.
Polymerization kinetics of reactions initiated by 4 µM eosin and 0.4 mM, 8 mM or 40 mM MDEA in the absence (A) and presence (B) of 0.4 mM TEMPO. Monomer contains 420 mM PEGDA and 35 mM VP in carbonate buffer. Carbonate buffer maintains pH = 9, despite changes in MDEA concentration. Samples were irradiated with 75 mW/cm2 >480 nm light.
Reports have previously been made regarding other reactions by which eosin or similar dye sensitizers for photoinitiation are able to regenerate. In one of these previously described reactions, two semi-reduced dye radicals react by disproportionation to regenerate one molecule of dye and one molecule of “leuco” or colorless dye.9,32 This reaction would act to prolong the time until all eosin is photobleached; however, because of the need to consume one eosin molecule for each that is regenerated, it cannot yield sufficient eosin regeneration to overcome the 100- to 1000-fold excess inhibitor as demonstrated here. As continued polymerization is observed even 15 or more minutes into the reaction (Figure 8B), it is concluded that although photobleaching reactions of eosin are certainly occurring, regeneration reactions of eosin are sufficiently greater in prevalence and rate to enable continued initiation over extended times.
In a second reported regeneration reaction of dye sensitizers, oxygen reacts with either semireduced dye radicals or leuco dye to regenerate the original dye.9,23 Clearly, this reaction would confer some amount of oxygen resistance to eosin-initiated polymerizations. Yet, the fact that eosin is relatively insensitive to TEMPO inhibitor even when the system has been bubbled with argon to reduce dissolved oxygen content, indicates that interaction with oxygen cannot be the only mechanism by which eosin regenerates. Figure 3A depicts 0.4 µM eosin initiating polymerization in the presence of 60 µM TEMPO. If regeneration of eosin radicals occurred only by reaction with molecular oxygen, then this system would require at least 60 µM oxygen to overcome 60 µM TEMPO. However, from Figure 4 it was estimated that the concentration of inhibitors (oxygen plus any others) in these systems prior to adding TEMPO is only 4 µM, much less than the 60 µM oxygen that would be necessary if eosin were regenerating by reaction with oxygen alone.
Notably, eosin is able to overcome 1000-fold excess oxygen, while it only overcomes 100-fold excess TEMPO. One important difference between oxygen and TEMPO inhibition is that in the first case eosin radicals may be regenerated by both molecular oxygen and by peroxy radicals, while in the case of TEMPO, eosin is only regenerated by reacting with TEMPO radicals prior to the inhibition event, not after TEMPO has already reacted with an MDEA radical or a propagating chain. The additional eosin regeneration pathways available in the presence of oxygen account for eosin’s greater ability to overcome oxygen inhibition as compared to TEMPO inhibition. In addition, the reactions of eosin radicals with molecular oxygen and/or peroxy radicals may be inherently faster than reaction of eosin radicals with TEMPO.
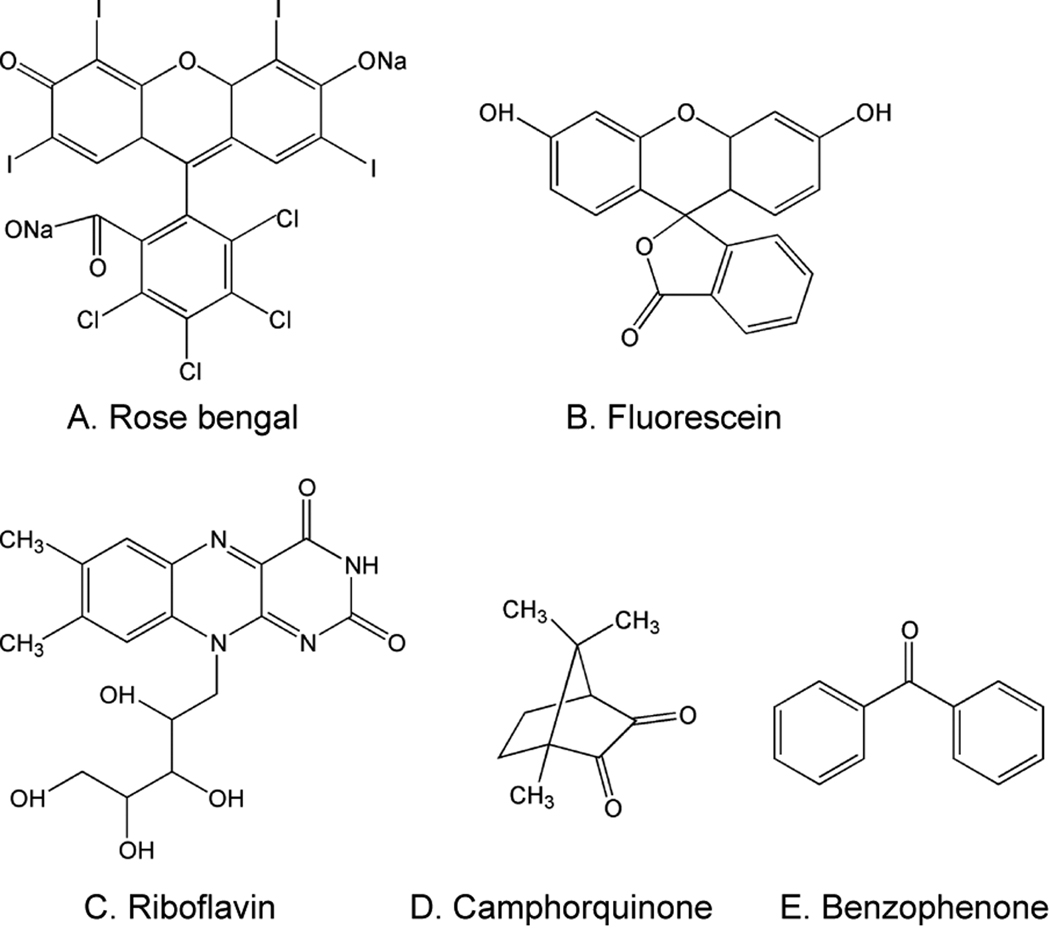
Finally, to investigate whether the proposed mechanisms may be applicable to additional type II photoinitiators, various photosensitizers were evaluated for their ability to initiate polymerization using MDEA as a coinitiator in the presence of TEMPO. Rose bengal and fluorescein (Figure 9), which, like eosin, are xanthene dyes, were able to overcome at least 100-fold and tenfold TEMPO, respectively (Table 3). Additionally, riboflavin, which is not a xanthene dye, initiated polymerization despite the presence of three fold excess TEMPO. Finally, neither camphorquinone nor benzophenone was able to overcome even equimolar concentrations of TEMPO. The fact that all three xanthenes dyes tested (eosin, rose bengal, and fluorescein) are capable of overcoming at least ten-fold excess TEMPO suggests that cyclic regeneration of the dye plays a significant role in the photoinitiation kinetics of xanthene dyes in general. Further, since riboflavin shows some capability to overcome TEMPO, there is evidence that cyclic regeneration contributes to the initiation kinetics of other classes of dye photosensitizers as well. In the case of camphorquinone and benzophenone, these data indicate that either cyclic regeneration of these dyes does not occur, or that it occurs much less readily than for these other initiation systems.
Figure 9.
Chemical structures for rose bengal (A), fluorescein (B), riboflavin (C), camphorquinone (D), and benzophenone (E).
Table 3.
aEffect of adding TEMPO to various type II photoinitiation systems.
| Initiator | TEMPO | TEMPO/ | Average Conversion (%) | |
|---|---|---|---|---|
| Initiator | (M) | (M) | Initiator | (14.5 min – 15.5 min) |
| Rose bengal | 4.0 × 10−6 | 0 | - | 65.2 (+/− 0.8) |
| 4.0 × 10−4 | 100 | 30.5 (+/− 0.4) | ||
| Fluorescein | 4.0 × 10−6 | 0 | - | 86.0 (+/− 1.2) |
| 4.0 × 10−5 | 10 | 30.5 (+/− 0.4) | ||
| Riboflavin | 3.6 × 10−5 | 0 | - | 31.2 (+/− 0.2) |
| 1.1 × 10−4 | 3 | 12.5 (+/− 0.2) | ||
| Camphorquinone | 4.0 × 10−4 | 0 | - | 20.7 (+/− 0.2) |
| 4.0 × 10−4 | 1 | 1.3 (+/− 0.2) | ||
| Benzophenone | 4.0 × 10−4 | 0 | - | 61.0 (+/− 1.3) |
| 4.0 × 10−4 | 1 | 0.2 (+/− 0.2) | ||
All polymerizations were performed with 420 mM PEGDA, 210 mM MDEA, and 35 mM VP in water. All samples were purged with argon for 20 minutes prior to polymerization. Rose bengal was exposed to 40 mW/cm2, >480 nm light. Fluorescein, riboflavin, and camphorquinone were exposed to 55 mW/cm2, 400–500 nm light. Benzophenone was exposed to 4 mW/cm2 320–390 nm light.
Conclusions
The data presented here highlight the fact that initiator selection has a strong impact on the inhibition sensitivity of photopolymerizations. Initiation of photopolymerization by eosin and MDEA is demonstrated to be an extremely effective method to alleviate the effects of oxygen and TEMPO inhibitors. It is anticipated that eosin will have similarly reduced sensitivity to other inhibitors that are either radicals themselves or generate radical intermediates during the inhibition process, a criteria that encompasses a wide variety of inhibitors. In addition to eosin, rose bengal, fluorescein, and riboflavin, many other dye photosensitizers, especially but not exclusively xanthene dyes, are expected to be likewise resistant to the effects of polymerization inhibitors.
Acknowledgements
This material is based upon work supported by NIH R21 CA 127884 and a National Science Foundation Graduate Research Fellowship to HJA. Also, this work has been supported by the State of Colorado and the University of Colorado Technology Transfer Office.
References
- 1.Bowman CN, Kloxin CJ. AIChE J. 2008;54:2775–2795. [Google Scholar]
- 2.Odian G. Principles of Polymerization. 4th Edition. New Jersey: John Wiley & Sons; 2004. 261, 326, 208, 214. [Google Scholar]
- 3.Becker H, Vogel H. Chem Eng Technol. 2006;29:1227–1231. [Google Scholar]
- 4.Kloosterboer JG. Adv Polym Sci. 1988;84:1–61. [Google Scholar]
- 5.Decker C, Jenkins AD. Macromolecules. 1985;18:1241–1244. [Google Scholar]
- 6.Anseth KS, Wang CW, Bowman CN. Macromolecules. 1994;27:650–655. [Google Scholar]
- 7.Roper TM, Kwee T, Lee TY, Guymon CA, Hoyle CE. Polymer. 2004;45:2921–2929. [Google Scholar]
- 8.White TJ, Liechty WB, Guymon CA. J Polym Sci Part A: Polym Chem. 2007;45:4062–4073. [Google Scholar]
- 9.Davidson RS. Chapter 5. In: Fouassier JP, Rabek JF, editors. Radiation Curing in Polymer Science and Technology. Vol. 3. New York: Elsevier Applied Science; 1993. pp. 153–176. [Google Scholar]
- 10.Hoyle CE, Lee TY, Roper T. J Polym Sci Part A: Polym Chem. 2004;42:5301–5338. [Google Scholar]
- 11.O’Brien AK, Cramer NB, Bowman CN. J Polym Sci Part A: Polym Chem. 2006;44:2007–2014. [Google Scholar]
- 12.Cramer NB, Bowman CN. J Polym Sci Part A: Polym Chem. 2001;39:3311–3319. [Google Scholar]
- 13.Cruise GM, Hegre OD, Scharp DS, Hubbell JA. Biotechnol Bioeng. 1998;57:655–665. doi: 10.1002/(sici)1097-0290(19980320)57:6<655::aid-bit3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama Y, Kameo T, Ohtaka A, Hirano Y. J Photochem Photobiol A. 2006;177:205–211. [Google Scholar]
- 15.Satoh M, Shirai K, Saitoh H, Yamauchi T, Tsubokawa N. J Polym Sci Part A: Polym Chem. 2004;43:600–606. [Google Scholar]
- 16.Blaya S, Murciano A, Acebal P, Carretero L, Ulibarrena M, Fimia A. Appl Phys Lett. 2004;84:4765–4767. [Google Scholar]
- 17.Hansen RR, Johnson LM, Bowman CN. Anal Biochem. 2009;386:285–287. doi: 10.1016/j.ab.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Avens HJ, Bowman CN. Acta Biomater. doi: 10.1016/j.actbio.2009.06.008. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Roz M, Lalevée J, Morlet-Savary F, Allonas X, Fouassier JP. J Polym Sci Part A: Polym Chem. 2008;46:7369–7375. [Google Scholar]
- 20.Sandholzer M, Schuster M, Varga F, Liska R, Slugovc C. J Polym Sci Part A: Polym Chem. 2008;46:3648–3661. [Google Scholar]
- 21.Popielarz R, Vogt O. J Polym Sci Part A: Polym Chem. 2008;46:3519–3532. [Google Scholar]
- 22.Lalevée J, El-Roz M, Allonas X, Fouassier JP. J Polym Sci Part A: Polym Chem. 2008;46:2008–2014. [Google Scholar]
- 23.Tigulla P, Vuruputuri U. J Chem Sci. 2004;116:115–118. [Google Scholar]
- 24.Avens HJ, Randle TJ, Bowman CN. Polymer. 2008;49:4762–4768. doi: 10.1016/j.polymer.2008.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen RR, Sikes HD, Bowman CN. Biomacromolecules. 2008;9:355–362. doi: 10.1021/bm700672z. [DOI] [PubMed] [Google Scholar]
- 26.Hansen RR, Avens HJ, Shenoy R, Bowman CN. Anal Bioanal Chem. 2008;392:167–175. doi: 10.1007/s00216-008-2259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gou L, Coretsopoulos CN, Scranton AB. J Polym Sci Part A: Polym Chem. 2004;42:1285–1292. [Google Scholar]
- 28.Green DW, Perry RH. Perry’s Chemical Engineers’ Handbook. 8th Edition. New York City: McGraw-Hill; 2008. Table 2–123. [Google Scholar]
- 29.Georges MK, Veregin RPN, Kazmaier PM, Hamer GK. Macromolecules. 1993;26:2987–2988. [Google Scholar]
- 30.Hawker CJ, Barclay GG, Orellana A, Dao J, Devonport W. Macromolecules. 1996;29:5245–5254. [Google Scholar]
- 31.Kizilel S, Pérez-Luna VH, Teymour F. Langmuir. 2004;20:8652–8658. doi: 10.1021/la0496744. [DOI] [PubMed] [Google Scholar]
- 32.Padon KS, Scranton AB. J Polym Sci Part A: Polym Chem. 2001;39:715–723. [Google Scholar]
- 33.Kharasch MS, Nudenberg W, Mantell GJ. J Org Chem. 1951;16:524–532. [Google Scholar]
- 34.Kaminsky V, Buback M, Egorov M. Macromol Theory Simul. 2002;11:128–135. [Google Scholar]