Abstract
Protein structures have proven to be a crucial piece of information for biomedical research. Of the millions of currently sequenced proteins only a small fraction is experimentally solved for structure and the only feasible way to bridge the gap between sequence and structure data is computational modeling. Half a century has passed since it was shown that the amino acid sequence of a protein determines its shape, but a method to translate the sequence code reliably into the 3D structure still remains to be developed. This review summarizes modern protein structure prediction techniques with the emphasis on comparative modeling, and describes the recent advances in methods for theoretical model quality assessment.
Keywords: protein structure prediction, CASP, comparative modeling, model quality assessment
In the late 1950s Anfinsen and co-workers suggested that an amino acid sequence carries all the information needed to guide protein folding into a specific spatial shape of a protein [1]. In turn, protein structure can provide important insights into understanding the biological role of protein molecules in cellular processes. Thus, in principle it should be possible to form a hypothesis concerning the function of a protein just from its amino acid sequence, using the structure of a protein as a stepping stone towards reaching this goal.
Modern experimental methods for determining protein structure through X-ray crystallography or NMR spectroscopy can solve only a small fraction of proteins sequenced by the large-scale genome sequencing projects, because of technology limitations and time constraints. Currently, there are more than 6,800,000 protein sequences accumulated in the non-redundant protein sequence database (NR; accessible through the National Center for Biotechnology Information: ftp://ftp.ncbi.nlm.nih.gov/blast/db/) and fewer than 50,000 protein structures in the Protein Data Bank (PDB; http://www.rcsb.org/pdb/). With these numbers at hand, it seems that the only way to bridge the ever growing gap between protein sequence and structure is computational structure modeling.
Computational protein structure prediction is a dynamic research field steadily increasing its outreach to biotechnology communities. Several reviews have been published recently on various aspects of structure modeling [2-14]. With the improvement of prediction techniques, protein models are becoming genuinely useful in biology and medicine, including drug design. There are numerous examples where in silico models were used to infer protein function, hint at protein interaction partners and binding site locations, design or improve novel enzymes or antibodies, create mutants to alter specific phenotypes, or explain phenotypes of existing mutations. What are the basic differences between the underlying methods, how can they be properly used, and which models can be trusted? The aim of this review is to address these issues from the standpoint of CASP (critical assessment of techniques for protein structure prediction) [15] with the focus placed on two practical issues – template-based modeling and assessment of model quality.
Protein structure modeling and CASP
Every other year since 1994, protein structure modelers from around the world dedicate their late spring and summer to testing their methods in the worldwide modeling experiment called CASP [15]. Predictors with expertise in applied mathematics, computer science, biology, physics and chemistry in well over one hundred scientific centers around the world work for approximately three months to generate structure models for the set of several tens of protein sequences selected by the experiment organizers. The proteins suggested for prediction (in the CASP jargon – “targets”) are either structures soon-to-be solved or structures already solved and deposited at the PDB but kept inaccessible to the general public until the end of the CASP prediction season. As soon as the predictions on a target are collected and the target structure itself is available, the Prediction Center at the University of California Davis performs an evaluation of the submitted models comparing them to the “gold standard” – the experimental structures. The results of evaluations [16] are made available to the independent assessors – experts in the field – who analyze the predictions first without knowing the identity of prediction groups, and present their analysis to the community at the predictors' meeting usually held in December of a CASP year. At that time, results of the evaluations are made publicly available through the Prediction Center website (http://predictioncenter.org) allowing predictors to compare their own models with those submitted by other groups. The papers by the assessors and by the most successful prediction groups are published in special issues of the journal Proteins: Structure, Function and Bioinformatics. There are now seven such issues available, one for each of the seven CASP experiments held so far. The details of the latest completed CASP, held in 2006, are described in the most recent CASP issue [15] and will be briefly discussed here. CASP8 is currently under way. In August 2008 the Prediction Center finished collecting structure predictions from 165 participating structure modeling groups, from 25 countries. More than 55,000 structure predictions were submitted for assessment. The CASP8 assessors are currently working on the analysis of the submitted predictions. The meeting to discuss the results takes place in Sardinia, Italy, in December 2008.
As defined by the current CASP classification, protein structure prediction methods can be divided into two broad categories: template-based modeling (TBM) and free modeling (FM). Each of these categories can be further split into two subcategories: TBM – into comparative modeling and fold recognition; and FM – into knowledge-based de novo modeling and ab initio modeling from first principles.
Template-based modeling in light of CASP
The template-based methods currently offer the most reliable prediction results but their applicability is limited to cases where it is possible to identify a structurally similar protein that can be used as a template for building the model.
Chothia and Lesk [17] have shown that protein structure is much more evolutionarily conserved than sequence and therefore similar sequences normally yield similar 3D structures. Based on this notion it is logical to check the target protein for sequence similarity to proteins of known structure and then to proceed with modeling starting from the structure of its homologue. The analysis of new, previously unknown or barely studied genomes indicates that new families are being discovered at a rate that is linear or almost linear with the addition of new sequences [18]. And, if at least a single structure within a family is determined experimentally, then homology modeling can be used to model the structures of all members of that family. Unless experimental methods undergo a major advance in the near future a major advancement allowing for a considerably faster and cheaper structure determination, homology modeling will remain a major player in many structure biology projects.
Even if a template for a modeled protein cannot be identified using sequence similarity, suitable templates may still exist among the already known structures, as Nature tolerates only a limited number of folds. The methods that check compatibility of a protein with existing folds using more sophisticated analysis (profile–profile sequence comparison, secondary structure comparison, knowledge-based potentials, etc.) may stand the chance of identifying the correct fold [2,19]. Using these methods it is, in many cases, possible to “thread” the sequence of the protein of unknown structure through the structure of an analogous protein, thereby using it as a template for modeling.
Historically, separate sets of modeling techniques were used in these two modeling difficulty cases. Owing to the recent rather impressive progress in the development of fold recognition methods [20,21], especially in their ability to detect remote homologues and produce better template-target alignments [2], the line separating the results obtained with the two classes of methods has blurred. Starting in CASP7 (2006), comparative modeling and fold recognition methods are assessed together in one template-based difficulty category.
Assessment of the template-based predictions in the latest completed CASP [22] identified the top two teams achieving particularly promising results: groups of Yang Zhang (University of Kansas) and David Baker (University of Washington). Both groups used highly automated computational approaches and, while Baker's group utilized hundreds of thousands of CPUs distributed worldwide to build the optimal model (http://boinc.bakerlab.org/rosetta/), Zhang's methodology necessitated considerably less CPU time. Zhang's approach is based on the improved I-TASSER methodology [23] that threads targets through the PDB library structures, uses continuous fragments in the alignments to assemble the global structure, fills in the unaligned regions by means of ab initio simulations and finally refines the assembly by an iterative lowest energy conformational search. Baker's template-based modeling uses three different strategies depending on the target length and target-template sequence similarity [24], and in general relies on computationally demanding sampling of conformational space coupled with an iterative all-atom refinement. The predictions from both groups improved over the best existing templates for the majority of template-based targets. Also, servers (fully automated predictors) from both of these groups performed well in CASP7, with Zhang-server having an edge over the Robetta server (from Baker's group) and placing third overall behind only the two already mentioned human-expert groups. The other top performing servers in CASP7 were those of Soeding et al. (HHpred2; BayesHH; HHpred3), Elofsson and co-workers (Pmodeller6), and Skolnick and co-workers (MetaTASSER) [25]. Analyzing progress of server performance in successive CASPs, it is evident that the gap between the best servers and the best human-expert groups is narrowing over time [21]. Especially in the case of easy template-based modeling, the progress of automated servers is impressive, with the fraction of targets where at least one server model is among the best six submitted models – increasing from 35% in CASP5 to 65% in CASP6, and to over 90% in CASP7. This also confirms the notion that the impact of human expertise on modeling of easy comparative targets is now marginal. In general, in CASP7 servers were at least on a par with humans (three or more models in the best six) for about 20% of targets; and significantly worse than the best human model for only very few targets. In CASP7 special attention was dedicated to the assessment of model details in high accuracy template-based modeling. The assessment showed that the group of Jooyoung Lee (Korea Institute for Advanced Study) generated models superior to those of other groups when compared, based on the overall structure quality, side-chain rotamer quality and suitability for crystallographic molecular replacement [26]. The approach of the Lee group is based on conventional backbone and side-chain modeling/refinement, multiple conformational space annealing and a score function consistency analysis. For most high accuracy template-based targets their models were closer to the native structure than any of the available template structures.
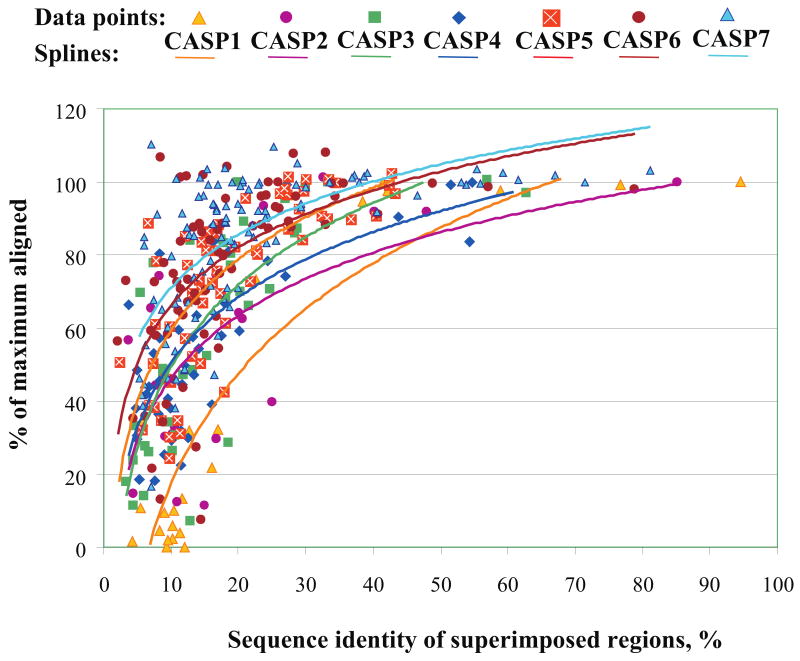
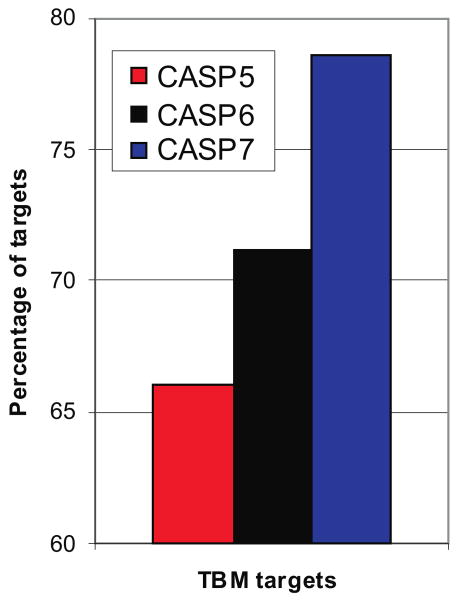
So, what are the main challenges still remaining in template-based modeling? Conceptually, the template-based modeling procedure starts from identifying and selecting the appropriate templates and follows with the target-template sequence alignment. And, after years of development, the level of target-template structural conservation and the accuracy of the alignment still remain the two issues having the major impact on the quality of resulting models. Figure 1 shows the alignment accuracy for all CASPs, as a percentage of the maximum template-target alignability. If the target-template sequence identity falls below the 20% level, as many as half of the residues in the model may be misaligned. There are several such examples even in the most recent CASP. At the same time, the best models for the majority of targets with greater than 30% sequence identity to a template have practically all residues correctly aligned. Closer inspection shows that in the latest two CASPs there were 14 targets at low sequence identity that have a higher fraction of the residues aligned than it is possible to achieve from a single template. This is encouraging, as these analyses show that modeling adds value over simply copying from best template structures [21,22,26]. Figure 2 shows that the percentage of targets where the best submitted models are better than a naïve single template model is steadily growing for the latest three CASPs. In CASP7, almost 80% of the best models in this category have registered added value over the naïve model. Model features not present in the best template may be added by refining aligned regions away from the template structure towards the experimental one, by using template free modeling methods and by adding features that are present in the other available templates [23,24,27-31].
FIGURE 1. Alignment accuracy for the best model of each target in all CASPs as percentage of maximum alignability.
Targets are ordered by sequence identity between the target and the closest template. Values on the vertical axis represent the ratio between the number of residues correctly aligned to the target structure in the best model and the maximum number of residues that can be aligned between the best available template and the target structure. An alignment of 100% indicates that all residues with an equivalent in the template were correctly aligned. A value greater than 100% indicates an improvement in model quality beyond that obtained by copying a template structure. Values above 100% may be obtained by using additional templates, through refinement or with template free methods. Trend lines show a steady though sometimes modest improvement over successive CASPs.
FIGURE 2. Fraction of targets for which the best submitted models in the template-based category are superior to the models naively built on the best template in CASPs 5–7.
Summarizing, currently available template-based methods can reliably generate accurate high-resolution models, comparable in quality to the structures solved by low resolution X-ray crystallography, when sequence similarity of a homologue to an already solved structure is high (50% or greater) [3,21,32]. As alignment problems are rare in these cases, the main focus shifts to accurate modeling of structurally variable regions (insertions and deletions relative to known homologues) and side chains, as well as to structure refinement. The high-quality comparative models often present a level of detail that is sufficient for drug design, detecting sites of protein–protein interactions, understanding enzyme reaction mechanisms, interpretation of disease-causing mutations and molecular replacement in solving crystal structures [7,11,32-34]. Even though there is a strong correlation between the target-template sequence similarity and the quality of the resulting model, the sequence similarity is not the only parameter determining the outcome of a modeling exercise, and in many cases high-resolution models can be built when sequence similarity is lower (30–50%). Typically, in this range medium resolution models are obtained, showing about 2–3 Ångstrom root mean square deviation (RMSD) from the native structure [3,32]. Even though less accurate, these models can also be used in many biological applications [11,32].
Modeling proteins of unknown fold
As the sequence similarity between the target and the available templates falls into the so-called twilight zone (sequence identity <25%), the resulting models become less accurate. In the most challenging cases, where neither significant sequence similarity nor structural similarity can be detected, the so-called free modeling methods are called upon [5,8,9]. CASP experiments demonstrate that the quality of free modeling predictions remains in general poor compared with predictions that are based on templates [20,21,35] and insufficient for many biomedical applications. At the same time these methods do occasionally produce quite accurate models for a few of the smaller targets. For example, one of the best groups in the free modeling category in recent CASPs, the Baker group (University of Washington) has their average CA-atom RMSD on free modeling targets around 12 Ångstroms (in CASP6 and 7). At the same time, in CASP6 this group obtained a close to high-resolution accuracy (<2 Ångstroms) in modeling of a 70-residue hypothetical protein from thermos thermophilus (T0281, PDB code - 1whz) by using structure assembly from fragments followed by extensive model refinement. In the next CASP, another impressive model was generated for a 97-residue protein (T0283, PDB code – 2hh6), where the model was again within 2 Ångstroms of the native structure over most of the chain. Recently, several notable cases of high-resolution structure prediction in the absence of a suitable structural template were reported in the literature [36-38]. The atomic accuracy of these predictions was then confirmed by X-ray crystallography. The above examples give hope for more practical applications of these techniques in the near future.
Model quality assessment (MQA)
Unlike experimentally derived structures, where accuracy of the structure can be estimated from experiment and typically falls within a narrow range, models are typically left un-annotated with quality estimates and can span a broad range of the accuracy spectrum.
Over the past two decades, a number of approaches have been developed to analyze correctness of protein structures and models. These methods use stereochemistry checks, molecular mechanics energy-based functions, statistical potentials, and machine learning approaches to tackle the problem. Typically, the features taken into account are the molecular environment, hydrogen bonding, secondary structure, solvent exposure, pair-wise residue interactions and molecular packing. Several detailed reviews of these techniques are available [39-41]. Recently, a rapid development of new methods in model quality assessment is taking place. The necessity of an unbiased evaluation of these methods led to their inclusion as a separate category in CASP, starting in 2006.
CASP7 assessment of MQA programs
More than 23,000 server-generated models of the 95 CASP7 targets were suggested for model quality assessment. The predictors were asked to submit a single global quality score for the entire model as well as assign error estimates on per-residue basis. At the end of the experiment, the observed model quality was compared with values submitted by predictors. CASP7 evaluation [42] demonstrates that at the moment the most consistent are the methods that rely on the availability of multiple models for the same protein (so-called consensus-based or clustering methods).
Two research groups, Pcons (Sweden) and LEE (Korea), outperformed other CASP7 participants in a statistically significant manner based on the results of the paired t-test assessment. Wallner and Elofsson's Pcons [43] is a consensus-based method capable of a quite reliable ranking of model sets for both easy and hard targets. Pcons uses a meta-server approach (i.e. combines results from several available well-established QA methods) to calculate a quality score reflecting the average similarity of a model to the model ensemble, under the assumption that recurring structural patterns are more likely to be correct than those observed only rarely. The LEE group, by contrast, based their technique on a comparison of query models with their own, and assigned rank in accordance with the distance between the models. Although both methods could provide a ranking significantly correlated with the one derived from CASP data, they were not able to select the best model consistently from the entire collection of models, indicating that considerable additional effort is needed in this area.
In addition to a good global quality performance, Pcons also showed promising results in assessing accuracy of the specific regions in a model, at times reproducing almost the exact C-alpha–C-alpha deviation along the sequence.
It should be underscored that, while the consensus-based methods are useful in model ranking, they can be biased by the composition of the set and, in principle, are incapable of assessing the quality of a single model.
Recent advances in MQA methods
In the two years following CASP7, a considerable increase in method development in the area of model quality assessment can be observed. More than a dozen papers have been published on the subject, and 45 quality assessment methods, almost double the CASP7 number, have been submitted for evaluation to CASP8. One way to classify quality assessment methods is to ask whether they provide a single “global” score for a structure model or give reliability estimates for structural regions on a per-residue basis. Although improvement in the first category is still very much needed, developments in prediction of local model quality are especially encouraging, allowing for a much more detailed view of the model, including its potential applications. Below, we focus on methods developed in the past two years.
Addressing local quality of models
Chen and Kihara examined the stability of the target-template alignments relative to the corresponding sets of suboptimal alignments and developed a model quality estimator based on these parameters, called SPAD [44]. The authors find excellent correlation to the resulting global and local level alignment errors and structure quality measures. This is not entirely surprising because alignment errors lie at the root level of any comparative modeling exercise. An additional advantage of using a systematic alignment analysis is that errors can be estimated at an early stage and therefore influence potential template and alignment choices. Another method, recently developed by Gao et al. and called FragQA [45], addresses a very similar problem by predicting a C-alpha RMSD between the aligned ungapped template fragments and the corresponding fragments in the native structure. This is done using support vector regression on features extracted from the alignment of the two sequences and those of the template structure. As the dependence of model quality on the correctness of alignment has been underscored by every CASP experiment held to date, these are certainly welcome developments. One potential way to extend both of these approaches, is to take into consideration alignments with all relevant templates at the same time, as information stemming from multiple sequence alignments is well known to improve recognition of homology in general. Some steps in this direction were made by the Fiser laboratory [31] as well as Chodanowski et al. [46], both placing emphasis on the structural assessment of alternative alignments. Finally, Fasnacht et al. [41], starting with an analysis of local structure similarity measures, make a valuable contribution with their comparative study of several statistical potentials and structural features with regard to their usefulness in local error prediction.
Combining evaluators
A number of recently developed techniques aim at optimally combining several independent model quality measures into one single score, reflecting the accuracy of the entire model or acting as a selector of near-native structures in the decoy sets of models. Correctly weighing the individual contributions included in the total score is a long-standing problem and now machine learning techniques are extensively applied to address it. It should probably be noted that all the approaches discussed in this section combine evaluators at the level of entire models. Eramian et al. [47] used support vector regression merging assessment scores from physics-based energy functions, statistical potentials (including the recently developed DOPE [48]) and machine-learning-based scoring functions to select the most accurate models from among a set of decoys (SVMod). They observe that the most significant contributions in the final score come from statistical (knowledge based) potentials, including surface accessibility and contact components. In other work from the same laboratory, Melo and Sali [39] use a genetic algorithm combining statistical potentials, stereochemical parameters, sequence alignment scores, geometrical descriptors and measures of protein packing to discriminate between models with correct and incorrect fold (GA_341). They identify statistical potentials incorporating contact and accessibility terms, as well as structure compactness, as features contributing the most to the final score. Also Benkert et al. [49] (QMEAN, regression analysis) and Merghetti et al. [50] (AIDE, neural networks) stress the importance of combining major geometrical aspects of protein structure, with the first work identifying an addition of a new three-residue torsion angle potential to the scoring function as particularly effective in differentiating between good and bad models. The contributions from Qiu et al. [51] and Zhou and Skolnick [52,53] differ by including the consensus-based features (i.e. incorporating in the analysis information from multiple models on the same target). While Zhou and Skolnick use a consensus C-alpha contact potential, Qiu et al. rely on support vector regression analysis to select the features most relevant to the final score (SVR). They identify consensus analysis as particularly important, with the corresponding weight almost ten times larger than the next one, corresponding to a pairwise atomic potential. Finally, Sadowski and Jones [54] and McGuffin [55,56], while placing emphasis on benchmarking individual methods, also offer neural network-based meta techniques that combine them. The first builds upon previously developed MODCHECK [57], while the second merges four original approaches in a program called ModFOLD. Assessing the relative performance of all the techniques discussed in this section is not a trivial matter, pointing perhaps to the significance of independent assessments such as CASP. Table 1 summarizes the parameters of the available model quality assessment methods. Ultimately, we want to emphasize efforts incorporating model quality assessment scores directly into modeling servers, including MODELLER [58] and HOMA [59] packages.
TABLE 1. Summary of different model quality assessment methods tested in CASP7 (2006) and published afterwards.
Overview of the quality assessment methods include: method name; method authors; reference to the method publication; type of input [single-model (S), multiple-model – consensus/clustering (C), single-model with a clustering version of the method available (S+C)]; approach [original (O) or meta, i.e. combining several methods or several measures (M)]; coordinate completeness [requires full atom model (FA), works with C-alpha only models (CA) or backbone-only models (BB), can take backbone coordinates and build sidechains internally using publicly available methods (BB+)]; method granularity [global (G) or local (L)]; dominant computational techniques and input information used; online accessibility [stand alone (S) or integrated (I)].
| Method | Authors | Publ | Single model/consens | Orig./meta | Full atom/backbone/C-alpha | Global/local | Computational techniques/information used | Online access |
|---|---|---|---|---|---|---|---|---|
| AIDE | Mereghetti et al. | [50] | S | M | FA/BB+ | G | Neural network combining 15 structural parameters | S |
| Chodanowski et al | [46] | S | M | FA | L | 15 predictors using different force fields | ||
| DOPE | Shen, Sali | [48] | S | O | FA | G | Statistical potential | I |
| SVMod. | Eramian et al | [47] | S | M | FA | G | SVM combining 24 individual assessment scores | I |
| FragQA | Gao et al | [45] | S | O | BB | GL | SVM combining sequence and structural features | |
| HOMA | Bhattacharya et al | [59] | S | M | FA | G | Structure analysis | I |
| GA_341 | Melo, Sali | [40] | S | M | FA | G | Genetic algorithm combining 21 features | I |
| MODCHECK | Sadowski, Jones | [54] | S+C | M | FA | G | Neural network combining 30 features including independent force fields | SI |
| ModFOLD | McGuffin | [55, 56] | S+C | M | BB | G | Neural network combining 6 scores (structural features and force fields) | SI |
| ProSA-web | Wiederstein et al | [60] | S | O | CA | GL | Statistical potential | S |
| Pcons | Wallner, Elofsson | [43] | C | O | BB | GL | Neural network based on structural features | SI |
| ProQ/ProQres | Wallner, Elofsson | [43] | S | O | FA | GL | Neural network based on structural features | SI |
| ProQprof | Wallner, Elofsson | [43] | S | O | BB | GL | Neural network based on sequence alignment | S |
| ProQlocal | Wallner, Elofsson | [43] | S | M | FA | L | Sequence and structural features | S |
| QMEAN | Benkert et al. | [49] | S+C | M | FA | GL | Combination of 5 statistical potentials | S |
| SPAD | Chen, Kihara | [44] | S | O | BB | GL | Stability of sequence alignment | |
| SVR | Qiu et al. | [51] | C | M | FA, BB+ | G | SVM combining 25 features (21 structural) | I |
| TASSER | Zhou, Skolnick | [52, 53] | C | O | CA | G | Structure features, statistical potentials | I |
Concluding remarks
Protein structure prediction is now undergoing a qualitative transition from a primarily academic pursuit to practical applications in medicine and biotechnology. Currently available template-based methods can generate models with the level of detail that, in cases of high homology, is sufficient for applications as demanding as drug design. In many cases high-resolution models can be built even when sequence similarity is relatively low (30–50%). Free modeling methods are not mature enough for routine biomedical applications, but the first instances of high-resolution structure prediction in the absence of templates have now been reported. Since models are not experimental structures determined with known accuracy but predictions, it is vital to present the user with the corresponding estimates of model quality. Much is being done in this area but further development of tools to assess model quality reliably is needed. The latest advances in structure prediction and assessment of model quality are to be evaluated by CASP8 in December 2008.
Acknowledgments
This work was supported by the NIH (LM7085 to KF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature
- 1.Sela M, et al. Reductive cleavage of disulfide bridges in ribonuclease. Science. 1957;125(3250):691–692. doi: 10.1126/science.125.3250.691. [DOI] [PubMed] [Google Scholar]
- 2.Ginalski K, et al. Practical lessons from protein structure prediction. Nucleic Acids Res. 2005;33(6):1874–1891. doi: 10.1093/nar/gki327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginalski K. Comparative modeling for protein structure prediction. Curr Opin Struct Biol. 2006;16(2):172–177. doi: 10.1016/j.sbi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Schueler-Furman O, et al. Progress in modeling of protein structures and interactions. Science. 2005;310(5748):638–642. doi: 10.1126/science.1112160. [DOI] [PubMed] [Google Scholar]
- 5.Bujnicki JM. Protein-structure prediction by recombination of fragments. Chembiochem. 2006;7(1):19–27. doi: 10.1002/cbic.200500235. [DOI] [PubMed] [Google Scholar]
- 6.Dunbrack RL., Jr Sequence comparison and protein structure prediction. Curr Opin Struct Biol. 2006;16(3):374–384. doi: 10.1016/j.sbi.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Nayeem A, et al. A comparative study of available software for high-accuracy homology modeling: from sequence alignments to structural models. Protein Sci. 2006;15(4):808–824. doi: 10.1110/ps.051892906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Floudas CA. Computational methods in protein structure prediction. Biotechnol Bioeng. 2007;97(2):207–213. doi: 10.1002/bit.21411. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol. 2008;18(3):342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moult J. Rigorous performance evaluation in protein structure modelling and implications for computational biology. Philos Trans R Soc Lond B Biol Sci. 2006;361(1467):453–458. doi: 10.1098/rstb.2005.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moult J. Comparative modeling in structural genomics. Structure. 2008;16(1):14–16. doi: 10.1016/j.str.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 13.Cozzetto D, et al. The evaluation of protein structure prediction results. Mol Biotechnol. 2008;39(1):1–8. doi: 10.1007/s12033-007-9023-6. [DOI] [PubMed] [Google Scholar]
- 14.Tramontano A, et al. The assessment of methods for protein structure prediction. Methods Mol Biol. 2008;413:43–57. doi: 10.1007/978-1-59745-574-9_2. [DOI] [PubMed] [Google Scholar]
- 15.Moult J, et al. Critical assessment of methods of protein structure prediction-Round VII. Proteins. 2007;69 8:3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kryshtafovych A, et al. New tools and expanded data analysis capabilities at the Protein Structure Prediction Center. Proteins. 2007;69 8:19–26. doi: 10.1002/prot.21653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. Embo J. 1986;5(4):823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5(3):e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu S, Zhang Y. MUSTER: Improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins. 2008;72(2):547–556. doi: 10.1002/prot.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kryshtafovych A, et al. Progress over the first decade of CASP experiments. Proteins. 2005;61 7:225–236. doi: 10.1002/prot.20740. [DOI] [PubMed] [Google Scholar]
- 21.Kryshtafovych A, et al. Progress from CASP6 to CASP7. Proteins. 2007;69 8:194–207. doi: 10.1002/prot.21769. [DOI] [PubMed] [Google Scholar]
- 22.Kopp J, et al. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69 8:38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69 8:108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 24.Das R, et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69 8:118–128. doi: 10.1002/prot.21636. [DOI] [PubMed] [Google Scholar]
- 25.Battey JN, et al. Automated server predictions in CASP7. Proteins. 2007;69 8:68–82. doi: 10.1002/prot.21761. [DOI] [PubMed] [Google Scholar]
- 26.Read RJ, Chavali G. Assessment of CASP7 predictions in the high accuracy template-based modeling category. Proteins. 2007;69 8:27–37. doi: 10.1002/prot.21662. [DOI] [PubMed] [Google Scholar]
- 27.Cheng J. A multi-template combination algorithm for protein comparative modeling. BMC Struct Biol. 2008;8:18. doi: 10.1186/1472-6807-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T, et al. Improving the accuracy of template-based predictions by mixing and matching between initial models. BMC Struct Biol. 2008;8:24. doi: 10.1186/1472-6807-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsson P, et al. Using multiple templates to improve quality of homology models in automated homology modeling. Protein Sci. 2008;17(6):990–1002. doi: 10.1110/ps.073344908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakravarty S, et al. Systematic analysis of the effect of multiple templates on the accuracy of comparative models of protein structure. BMC Struct Biol. 2008;8:31. doi: 10.1186/1472-6807-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Fuentes N, et al. Comparative protein structure modeling by combining multiple templates and optimizing sequence-to-structure alignments. Bioinformatics. 2007;23(19):2558–2565. doi: 10.1093/bioinformatics/btm377. [DOI] [PubMed] [Google Scholar]
- 32.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294(5540):93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 33.Raimondo D, et al. Automatic procedure for using models of proteins in molecular replacement. Proteins. 2007;66(3):689–696. doi: 10.1002/prot.21225. [DOI] [PubMed] [Google Scholar]
- 34.Kopp J, Schwede T. Automated protein structure homology modeling: a progress report. Pharmacogenomics. 2004;5(4):405–416. doi: 10.1517/14622416.5.4.405. [DOI] [PubMed] [Google Scholar]
- 35.Jauch R, et al. Assessment of CASP7 structure predictions for template free targets. Proteins. 2007;69 8:57–67. doi: 10.1002/prot.21771. [DOI] [PubMed] [Google Scholar]
- 36.Bradley P, et al. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309(5742):1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 37.Qian B, et al. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450(7167):259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang L, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319(5868):1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marti-Renom MA, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 40.Melo F, Sali A. Fold assessment for comparative protein structure modeling. Protein Sci. 2007;16(11):2412–2426. doi: 10.1110/ps.072895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fasnacht M, et al. Local quality assessment in homology models using statistical potentials and support vector machines. Protein Sci. 2007;16(8):1557–1568. doi: 10.1110/ps.072856307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cozzetto D, et al. Assessment of predictions in the model quality assessment category. Proteins. 2007;69 8:175–183. doi: 10.1002/prot.21669. [DOI] [PubMed] [Google Scholar]
- 43.Wallner B, Elofsson A. Prediction of global and local model quality in CASP7 using Pcons and ProQ. Proteins. 2007;69 8:184–193. doi: 10.1002/prot.21774. [DOI] [PubMed] [Google Scholar]
- 44.Chen H, Kihara D. Estimating quality of template-based protein models by alignment stability. Proteins. 2008;71(3):1255–1274. doi: 10.1002/prot.21819. [DOI] [PubMed] [Google Scholar]
- 45.Gao X, et al. FragQA: predicting local fragment quality of a sequence-structure alignment. Genome Inform. 2007;19:27–39. [PubMed] [Google Scholar]
- 46.Chodanowski P, et al. Local alignment refinement using structural assessment. PLoS ONE. 2008;3(7):e2645. doi: 10.1371/journal.pone.0002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eramian D, et al. A composite score for predicting errors in protein structure models. Protein Sci. 2006;15(7):1653–1666. doi: 10.1110/ps.062095806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15(11):2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benkert P, et al. QMEAN: A comprehensive scoring function for model quality assessment. Proteins. 2008;71(1):261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- 50.Mereghetti P, et al. Validation of protein models by a neural network approach. BMC Bioinformatics. 2008;9:66. doi: 10.1186/1471-2105-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu J, et al. Ranking predicted protein structures with support vector regression. Proteins. 2008;71(3):1175–1182. doi: 10.1002/prot.21809. [DOI] [PubMed] [Google Scholar]
- 52.Zhou H, Skolnick J. Ab initio protein structure prediction using chunk-TASSER. Biophys J. 2007;93(5):1510–1518. doi: 10.1529/biophysj.107.109959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou H, Skolnick J. Protein model quality assessment prediction by combining fragment comparisons and a consensus C(alpha) contact potential. Proteins. 2008;71(3):1211–1218. doi: 10.1002/prot.21813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadowski MI, Jones DT. Benchmarking template selection and model quality assessment for high-resolution comparative modeling. Proteins. 2007;69(3):476–485. doi: 10.1002/prot.21531. [DOI] [PubMed] [Google Scholar]
- 55.McGuffin LJ. The ModFOLD server for the quality assessment of protein structural models. Bioinformatics. 2008;24(4):586–587. doi: 10.1093/bioinformatics/btn014. [DOI] [PubMed] [Google Scholar]
- 56.McGuffin LJ. Benchmarking consensus model quality assessment for protein fold recognition. BMC Bioinformatics. 2007;8:345. doi: 10.1186/1471-2105-8-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettitt CS, et al. Improving sequence-based fold recognition by using 3D model quality assessment. Bioinformatics. 2005;21(17):3509–3515. doi: 10.1093/bioinformatics/bti540. [DOI] [PubMed] [Google Scholar]
- 58.Eswar N, et al. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 59.Bhattacharya A, et al. Assessing model accuracy using the homology modeling automatically software. Proteins. 2008;70(1):105–118. doi: 10.1002/prot.21466. [DOI] [PubMed] [Google Scholar]
- 60.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–410. doi: 10.1093/nar/gkm290. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]