Abstract
Two recent, large GWAS in European populations have associated a ∼47 Kb region that contains part of the FTO gene with high BMI. The functions of FTO and adjacent FTM in human biology are not clear. We examined expression of these genes in organs of mice segregating for monogenic obesity mutations, exposed to under/over feeding, and to 4 °C. Fto/Ftm expression was reduced in mesenteric adipose tissue of mice segregating for the Ay, Lepob, Leprdb, Cpefat or tub mutations and there was a similar trend in other tissues. These effects were not due to adiposity per se. Hypothalamic Fto and Ftm expression were decreased by fasting in lean and obese animals and by cold exposure in lean mice. The fact that responses of Fto and Ftm expression to these manipulations were almost indistinguishable suggested that the genes might be co-regulated. The putative overlapping regulatory region contains at least 2 canonical CUTL1 binding sites. One of these nominal CUTL1 sites includes rs8050136, a SNP associated with high body mass. The A allele of rs8050136 – associated with lower body mass than the C allele – preferentially bound CUTL1 in human fibroblast DNA. 70% knockdown of CUTL1 expression in human fibroblasts decreased FTO and FTM expression by 90 and 65 %, respectively. Animals and humans with various genetic interruptions of FTO or FTM have phenotypes reminiscent of aspects of the Bardet-Biedl obesity syndrome, a confirmed “ciliopathy”. FTM has recently been shown to be a ciliary basal body protein.
Keywords: obesity, hypothalamus, adipose tissue, CUTL1
Introduction
Heritability of adiposity –which reflects genetic contribution to the phenotype within a specific environment– is high, and variously estimated at 40-60% (28, 49). The search for the underlying genes for obesity –using conventional linkage, association and candidate gene approaches– has generated a large number of positive findings, many of which have not been replicated (e.g. 19, 25, 32, 37, 55, 67). Among the reasons for lack of consistent replication may be the relatively small population sizes, few markers genotyped, and blunt phenotypes. The recent generation of high density single nucleotide polymorphism (SNP) and haplotype maps (International HapMap project; http://www.hapmap.org/) has revolutionized the field of human quantitative genetics. Applied to large, suitably phenotyped groups of subjects, whole-genome association studies (GWAS) are implicating novel genes not previously considered based on extant understanding of the molecular physiology of specific phenotypes. The discovery of the “Fat Mass and Obesity Associated gene” (FTO) as a potentially important contributor to human adiposity is such an example.
In two GWAS involving a total of ∼42,000 obese and non-obese subjects, dose-dependent highly significant effects of specific SNPs on Chr. 16 have been associated with increased body mass index (BMI) (14, 52). In agreement with these results, Dina et al (8) identified an association between rs1121980 and morbid obesity (BMI≥40 Kg/m2) in 8,000 individuals of European ancestry, and replicated their finding in another cohort of 4,864 obese and non-obese subjects. Follow-up studies of relatively smaller cohorts confirmed association of the same region with obesity in German and Belgian children and adults (21, 46). However, no significant association was detected in Chinese and Oceanic populations (27, 44). The interval containing the associated SNPs spans ∼30 Kb and extends by linkage disequilibrium (LD) to a ∼47 Kb region containing parts of introns 1 and 2 and exon 2 of FTO (Fig. 1). The molecular function(s) of FTO, and the mechanism(s) by which these allelic variations convey effects on adiposity are not clear.
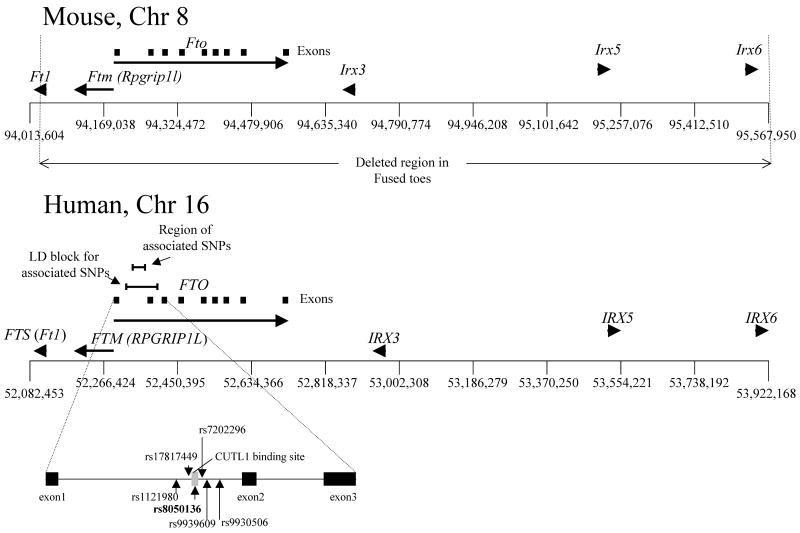
Figure 1. Organization of the Fto/Ftm gene cluster.
Positions of Fto and Ftm in mouse and human genomes. SNPs associated with BMI are indicated.
The transcriptional start of RPGRIP1L (human ortholog of mouse Ftm in the human; here referred to as FTM) is ∼3.4 kb upstream of FTO in humans. (Fig 1). By virtue of their close proximity to SNPs strongly associated with adiposity (e.g. rs9939609; 13), FTO or FTM, or both, may account for the association of this genetic interval with differences of adiposity in humans.
Fto was originally cloned in the mouse (48) and is part of a contiguous gene deletion in murine Fused toes (64) which has a 1.6 Mb deletion of Chr. 8 also containing Ftm, Ft1 and the Iroquois B cluster consisting of Irx3,5 and 6 (47) (Fig. 1). Homozygous mutants are embryonic lethal and display neural tube defects, left-right asymmetry (16, 20) and polydactyly (17). Embryos homozygous for the Fused toes deletion also have defects in both anteroposterior and dorsoventral patterning of the brain, including a reduction in the size of the hypothalamus that could be due to effects on Sonic Hedgehog (SHH)-related pathways (2). Heterozygous mice are not obese but have fused digits as well as hyperplasia of the thymus, possibly due to apparent impairment of programmed cell death (64). In humans, a de novo duplication of the region on chromosome 16q12.2 that includes FTO, FTM, FT1, RBL2 (retinoblastoma-like2) and NET1 [Solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2] is associated with anisomastia, somatic dysmorphisms, mental retardation and obesity (56).
Left-right asymmetry, neural tube patterning, and floor plate defects in the Fused toes mutant may be caused by the absence of Ftm, a regulator of SHH signaling expressed at the basal body of cilia (65). Adult mice lacking cilia throughout the central nervous system and, more specifically, pro-opiomelanocortin (POMC) neurons display increased food intake that leads to obesity (11). Ftm is homologous to RPGRIP1 (RPGR-interacting protein 1). Mutations of RPGRIP1 result in retinitis pigmentosa in humans (36) due to degeneration of photoreceptor cells possibly resulting from dysfunction of retinal cilia. (22, 45). In Bardet-Biedl syndrome, a syndromic form of human obesity associated with polydactyly, retinal degeneration and renal malformations, derangements of ciliary function have been implicated (39).
To further evaluate whether FTO or FTM might be responsible for the strong association of this genetic region with human adiposity, and to assess the molecular physiology of an implicated SNP (rs8050136) found within a putative transcription factor binding site (Fig. 1), we analyzed the expression of Fto and Ftm in several mouse models of obesity, and evaluated a possible functional role for this SNP as part of a putative Cutl-like 1 (CUTL1; transcription factor) binding site regulating FTO/FTM expression. Our intention was to identify differences of FTO/FTM expression in individual organs and specific fat depots upon environmental manipulations and in response to genetic disturbances of specific pathways (29) related to energy homeostasis.
Experimental Procedures
Mouse strains
Lepob (B6.V-Lepob/J), Leprdb (B6.Cg-m +/+ Leprdb/J), Cpefat [B6.HRS(BKS)-Cpefat/J], tub (B6(Cg)-Tubtub/J), Ay (B6.Cg-Ay/J), and control +/+ (C57BL/6J) male mice were obtained from The Jackson Laboratory (Maine, USA) at 4 weeks of age and sacrificed within 1 day of their arrival. At the Jackson Laboratory, and our laboratory, mice were fed regular chow (6% Kcal from fat: “NIH 31 6%”; Purina Mills, USA). C57BL/6J male diet-induced obese (DIO) mice were raised at the Jackson Laboratory. At 4 weeks of age, DIO mice were fed chow containing 10% Kcal from fat (Cat No. D12450B1; Open Source Diets™, USA) for 2 weeks, and then switched to high-fat chow (60% Kcal from fat; Cat No. D12492; Open Source Diets™, USA) for an additional 12 weeks. DIO control mice were fed chow containing 10% Kcal from fat (Cat No. D12450B1; Open Source Diets™, USA) at 4 weeks of age for a period of 14 weeks at the Jackson Laboratory. DIO and DIO control mice were sacrificed at 18 weeks of age, upon arrival. All mutant mice were sacrificed at 4 weeks of age in order to minimize possible secondary effects of obesity on Fto/Ftm expression.
Fasted mice were not fed for 40 hours. For the thermal challenge experiments, mice were placed singly in a 4 °C cold room for 30 min with ad libitum access to food and water.
Room temperature was constant at 21°C (unless otherwise stated) on a 12 h light/12 h dark cycle (lights were turned off at 7 pm). Mice had ad libitum access to food and water. All protocols were approved by the Columbia University Institutional Animal Care and Use Committee.
Body mass and composition measurements
To examine possible secondary effects of body composition on Fto/Ftm gene expression, body composition of the mice used in these studies was determined by TD-NMR using a Minispec Analyst AD lean fat analyzer (Bruker Optics, Silberstreifen Germany; the TD-NMR was calibrated according to the manufacturer's directions. The phenotypes were comparable to those reported in the literature (Table 1). In mice fasted for 40h, there were anticipated differences in responses of body weight and composition. Wild type C57BL/6J mice lost ∼50% of their total fat mass (p<0.02). Fasted Lepob mice lost from 5-15% of their total fat mass (p<0.05). Both Lepob and +/+ mice lost ∼20% of their total lean mass during the 40h period of food restriction (p<0.01).
TABLE 1. Body composition.
Fat and lean mass and fractional fat content of Lepob, Leprdb, Cpefat, tub, and +/+ (C57BL/6J) mice measured by TD-NMR. Body composition data for the Diet Induced Obese (DIO) mice and their controls were obtained from The Jackson Laboratory (Maine, USA; personal communication).
| Mouse (N=5) | Age (weeks) | Body Weight (g) [SD] | Fat (g) [SD] | Lean mass (g) [SD] | % fat [SD] |
|---|---|---|---|---|---|
| Lepob | 4 | 24.2 [0.9] | 9.4 [0.8] | 13.1 [0.3] | 39 [2.1] |
| Leprdb | 4 | 24.5 [2.2] | 8.5 [1.3] | 14.1 [0.8] | 35 [2.7] |
| tub | 4 | 27.0 [0.8] | 2.8 [0.3] | 22.8 [0.9] | 10 [1.2] |
| Cpefat | 4 | 20.5 [1.9] | 2.8 [0.5] | 17.2 [2.1] | 13 [1.4] |
| +/+ | 4 | 18.0 [1.3] | 2.1 [0.5] | 16.6 [1.5] | 11 [0.9] |
| DIO | 18 | 37.0 [2.9] | 12.9 [0.91] | 21.1 [1.7] | 35 [2.4] |
| DIO control | 18 | 30.0 [2.0] | 7.0 [0.7] | 2.01 [1.0] | 23 [2.2] |
Values are mean ± SD
Isolation of total RNA and cDNA synthesis
All mice were sacrificed between 2-4 PM. All tissues were dissected and immediately flash frozen in liquid N2. Each tissue was crushed into a fine powder in a 1.5ml eppendorf tube in the presence of liquid N2 and subsequently lysed in 1ml of Qiazol (Qiagen, Valencia, CA). Total RNA was extracted and DNase-treated using the RNeasy® Lipid Tissue Mini Kit and RNase-free DNase (Qiagen, Valencia, CA) according to the manufacturer's instructions. cDNA synthesis from 2.8 μg of total RNA was performed at 42°C for 55 min utilizing the Sprint™PowerScript™ PrePrimed Single Shots kit (CLONTECH, USA) with Oligo (dT)18 primers. Subsequently, the reverse transcriptase was inactivated at 72°C for 10 min.
qPCR
Transcript quantitation was performed in a Lightcycler 2.0 (ROCHE, USA) using the LightCycler ® FastStart DNA Master SYBR Green I kit (ROCHE, USA) according to the manufacturer's instructions. Samples were heated at 95°C for 10 min, followed by 40 cycles of 95 °C for 15 sec, 60-65°C for 10 sec, and 72 °C for 12 sec. For the amplification of the endogenous control genes Gapdh and Rps3, the annealing temperature was 60 °C and 58 °C, respectively. The annealing temperature was 65 °C for the primer pairs specific to the remaining genes. Each 20 μl reaction contained 2.5 μl of cDNA previously diluted to 1/11, 1.6 mM MgCl2, 3 micromolar of each primer and the recommended amount of the SYBR Green I mix. Primers for the endogenous control genes used were as follows:
Gapdh
5′-GCAGTGGCAAAGTGGAGATTGTTGC
5′-CCCGTTGATGACAAGCTTCCCATTC,
Actb
5′-TCTGGTGGTACCACCATGTACCCAG
5′-TGGAAGGTGGACAGTGAGGCCAG,
Rps3
5′-ATCAGAGAGTTGACCGCAGTTG
5′-AATGAACCGAAGCACACCATAG
Hprt
5′- CGCAGTCCCAGCGTCGTGATTTAGC
5′- CCCATCTCCTTCATGACATCTCGAGC
The Fto primers amplified part of exons 2 (5′-CACTTGGCTTCCTTACCTGACCCCC) and 3 (5′-GGTATGCTGCCGGCCTCTCGG).
The Ftm primers amplified part of exons 4 (5′- CCAAACAGCAGCTCCAAGTCCAGGG) and exon 5 (5′- GAGCGTGGGTTGTACAGTTTCTGCTTC).
Transcript quantitation was performed using the automated absolute quantification mode of the LightCycler® Software. Standard curves were calculated for each set of primers using hypothalamic cDNA from +/+ controls. Both Fto and Ftm-specific primer sets had amplification efficiencies of 1.8. The crossing point was defined as the first maximum of the second derivative of the fluorescence curve.
Expression levels of Gapdh, Actb, Hprt and Rps3 were measured in subcutaneous, mesenteric, perirenal, epididymal fat, brown adipose tissue, pancreas, liver and hypothalamus of all mice used in these studies. Although expression levels of the same gene varied across tissues, transcript levels of Gapdh. Actb and Hprt showed only modest variation (1-5%) within the same tissue among DIO mice, or those segregating for the various mutations. In contrast, Rps3 expression was ∼70% lower (p<0.001) in mesenteric fat of Lepob fasted mice. We used Gapdh as a loading control for the expression analysis.
Alternative transcript analysis
Fto contains 9 exons (all coding). To enable analysis of organ-specific expression rates, we identified exons present in all or most mRNA isoforms. Total RNA from the hypothalamus and mesenteric fat of +/+ mice was extracted as described above. cDNA was synthesized using the Superscript™ III First-Strand Synthesis System for RT-PCR with oligo(dT)20 (Invitrogen, Carlsbad, California, USA). Forward and reverse primers were as follows:
Fto
exon1 forward (5′-GGCGAAGGCGGCTTTAGTAGCAG),
exon2 forward (5′-CACTTGGCTTCCTTACCTGACCCC),
exon 2 reverse (5′-GGGGTCAGGTAAGGAAGCCAAGTG),
exon 3 forward (5′- GCCACTGTGCATGGCAGAGTTCCCC),
exon 3 reverse (5′- GGGGAACTCTGCCATGCACAGTGGC),
exon 4 forward (5′- CAGGATTAACAATCCCTCTTCACCAGGG),
exon 4 reverse (5′-CCCTGGTGAAGAGGGATTGTTAATCCTG),
exon 5 forward (5′-CGGTTTAGTTCCACTCACCGTGTGG),
exon 5 reverse (5′ CCACACGGTGAGTGGAACTAAACCG),
exon 6 forward (5′- CTCAGACGATGGCGACGTCTCGTTG),
exon 6 reverse (5′- CAACGAGACGTCGCCATCGTCTGAG),
exon 7 forward (5′- TGGTGTGAGCCCATGACTCACCTGG),
exon 7 reverse (5′- CCAGGTGAGTCATGGGCTCACACCA),
exon 8 forward (5′-ACCGTGCGCCAGAACCTGAGGAAGG),
exon 8 reverse (5′- ACCGTGCGCCAGAACCTGAGGAAGG),
exon 9 reverse (5′- GGGCAGAGGCATGGAAGGGTCATCC).
Amplification of hypothalamic and mesenteric cDNA using all possible primer combinations was achieved by varying extension time from 30 sec to 10 min and annealing temperature between 65-68 °C. The resulting PCR products were sequenced bidirectionally by GENEWIZ (San Diego, California). We did not identify alternatively spliced species in hypothalamus or mesenteric fat. We also examined Fto cDNA entries in NCBI. Most Fto cDNA clones from mouse included exons 2 and 3 (GenBank accession numbers: AK049502, AJ237917, AK088881, AK040866, AK045465, BC057008, AK022222, AK161060, AK036677). Fto cDNA sequence entries that did not include exons 2 and 3 were cloned from human testis (Accession number AK016860) or human melanoma and melanocyte cells (Accession numbers AK210708, AK194211, AK184948). qPCR using the primers listed above showed no differences in transcript abundance for mRNA species listed in GenBank (data not shown). Primers that amplified the junction between exons 2 and 3 were used for the quantitative expression analyses reported here.
Ftm contains 26 exons (25 coding). Inspection of mouse and human entries in NCBI was used to identify isoforms present in the majority of transcripts. Two transcripts included exons 4 and 5 [GenBank accession numbers AK029911 (mouse testis) and AJ344253 (11-day old embryo)]. Shorter transcripts cloned from human testis, bone and 15-day mouse embryos lacked exons 4 –5, but did not contain exons in common among them (AK00675, AK053090, AK036554). qPCR showed no difference in Ftm transcript abundance in all mouse tissues tested (same as tested for Fto) among all mRNA species recorded in NCBI (data not shown). The primers used were as follows:
exon 4 forward 5′-CCAAACAGCAGCTCCAAGTCCAGGG,
exon 5 reverse 5′-GAGCGTGGGTTGTACAGTTTCTGCTTC,
exon 6 forward 5′-CTTCTTCAGCTTCGAGAACAGCAGGC,
exon 7 reverse 5′-GACTGAAAGAGCATTGCTTTTCTCC,
exon 10 reverse 5′-GAATTAATGATTTAGAAAAGGAGCGGGAAC.
We chose to amplify the junction between exons 4 and 5 in quantitative assessments of Ftm expression.
In situ hybridization
4-week old C57BL/6J or Lepob male mice were perfused with 4% paraformaldehyde, and 13.5 dpc old +/+ or Lepob embryos were fixed overnight in 4% paraformaldehyde. Tissues were excised, dehydrated in 30% sucrose, frozen, and placed on slides as 10 micron medial coronal hypothalamic, sagittal pancreatic or medial sagittal embryonic sections.
Probes for in situ hybridization of embryonic and adult tissue were made by amplifying a 792 bp fragment from hypothalamic cDNA of C57BL/6J mice spanning Fto exons 2-4:
5′-CACTTGGCTTCCTTACCTGACCCC,
5′- CCCTGGTGAAGAGGGATTGTTAATCCTG
and a 736 bp fragment spanning Ftm exons 2-6:
5′-GAGACCTGCCGGTGAAAGATACAGG,
5′- GCCTGCTGTTCTCGAAGCTGAAGAAG.
Each DNA fragment was cloned in the Dual Promoter pCR® II-TOPO® Vector provided with the TOPO TA Cloning® Kit (Invitrogen, Carlsbad, California, USA) according to the manufacturer's specifications. Sense and antisense riboprobes labeled with Digoxigenin (DIG; Roche, USA) were prepared by in vitro transcription using T7 or Sp6 RNA polymerase (Promega, USA) at 42°C for 180 min. In situ hybridization was performed as previously described (7).
Chromatin immunoprecipitation assay
Primary fibroblast cells (∼4×106) from human skin heterozygous for rs8050136 (A/C), and rs17817449 (T/G) (Fig. 1) were fixed with formaldehyde, lysed and sonicated using the Chromatin Immunoprecipitation (ChIP) Assay Kit (Millipore, USA) according to the manufacturer's specifications. Halt™ Protease and Phosphatase Inhibitor cocktails (PIERCE, USA) were used according to the manufacturer's specifications. Protein-DNA complexes were incubated with mouse CUTL1 (ABR, USA), mouse HES1 (Santa Cruz Biotechnology, Santa Cruz) or mouse IgG (ABR, USA) antibodies at a 1:400 dilution, and incubated with Protein A agarose beads in the presence of salmon sperm DNA. CUTL1, HES1, or IgG-DNA complexes were reverse cross-linked, and the unbound protein was digested following the protocol provided with the Chromatin Immunoprecipitation (ChIP) Assay Kit (Millipore, USA). The DNA fragments were purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA). Transcript detection was performed by qPCR in Lightcycler 2.0 (ROCHE, USA) using the LightCycler ® FastStart DNA Master SYBR Green I kit (ROCHE, USA) according to the manufacturer's instructions. Samples were heated at 95°C for 10 min, followed by 35 cycles of 95 °C for 15 sec, 55°C for 10 sec, 72 °C for 12 sec. Each 20 μl reaction contained 1 μl of ChIP DNA fragments, 1.6 mM MgCl2, 3 micromolar of each primer and the recommended amount of the SYBR Green I mix. The following primers were used:
rs8050136:
5′-CGGTATTTGATTTCCTTTTCCCTGGG,
5′-biotin-GCATTCCATGAGTCCATCTCTACAG,
rs17817449:
5′-GTAACTGAGTCTCCCCTAACTGG,
5′-biotin-GGGAGTGCACCAAATTCAAACC.
The Institutional Review Board at Columbia University Medical Center (USA) approved the study and the participant gave written informed consent.
Pyrosequencing
Pyrosequencing was performed as previously described (35). In place of magnetic streptavidin beads, streptavidin SepharoseTM High Performance (Amersham Biosciences AB, USA) beads were used to capture biotin-labeled DNA fragments. The following sequencing primers were used:
rs17817449: 5′-TCAGCTTGGCACACAGAAAC,
rs8050136: 5′-CCAGTTGCCCACTGTGGCA.
The nucleotide dispensing order for sequences [T/G]GTTTTAATT (rs17817449) and AT[C/A]AATAT (rs8050136) were C-T-G-C-T-A-T and C-A-T-C-A-G-T-A-T, respectively.
siRNA for CUTL1
Lipofectamine™ 2000 (Invitrogen, Carlsbad, California, USA) was used according to the manufacturer's specifications to transfect siRNA into primary fibroblasts (at ∼50% confluency) from human skin heterozygous for rs8050136 and rs17817449. For CUTL1, the siRNA target sequence and scrambled control were:
5′-GGCUGACUAUGAAGAGGUGAAGAAA and
5′-GGCUCUAUGAAGAGGGAGUAAGAAA,
respectively (Invitrogen, Carlsbad, California). Cells were harvested 48 hours later and total RNA isolated as previously described. The same qPCR protocols for Fto and Ftm were used to amplify comparable regions of the human orthologs:
FTO exons 2-3
5′-CACTTGGCTCCCTTATCTGACCCCC,
5′-GATACACTGCTGGCTTCTCGG
FTM exons 4-5
5′-CCAAACAGCAACTTCAAACCCAGGG,
5′-GTAAACATGGGATGTGGAGTTTCTGCTAC.
The same protocol was followed for the assessment of CUTL1 expression. The following CUTL1-specific primers were used to amplify part of exon 18 and 20 and the whole of exon 19:
5′- CTACATGTACCAGGAGGTGGACACCATCG,
5′- CTGCTGCAGCGGGTCCTGGAGCGAT.
Statistical analysis
Group data are expressed as means +/- standard deviation. Statistical analyses were performed using ANOVA, ANCOVA (StatView 5.0, SAS Institute Inc. or STATISTICA v. 6; StatSoft® Tulsa, Oklahoma). Data were corrected for repeated measures using repeated measures ANOVA (STATISTICA v. 6; StatSoft® Tulsa, Oklahoma). Levels of statistical significance were set at 2-tailed palpha<0.05. In all figures, error bars are SD.
Results
Comparison of Fto and Ftm expression in various tissues of C57BL/6J (+/+) mice
Fto transcript levels were higher than Ftm in all tissues tested (Fig 2). Fto expression was ∼6-fold higher than Ftm in the hypothalamus (p<0.001). Expression of Fto in the hypothalamus was ∼2-fold (p<0.001) and ∼25-fold higher (p<0.001) than mesenteric fat and brown adipose tissue, respectively. Expression levels of Ftm in the hypothalamus and mesenteric fat were at least ∼2-fold higher (p<0.01) than all other fat depots and the liver. Fto expression in the pancreas was ∼2-fold higher (p<0.02) than Ftm expression. By in situ hybridization, Fto was highly expressed throughout the hypothalamus, but relatively higher in the arcuate nucleus (Fig. 3A) while Ftm expression was restricted to the arcuate nucleus. These findings are consistent with comparable data available in the Allen Brain Atlas (http://www.allenbrainatlas.com). In the pancreas, by in situ hybridization, both Fto and Ftm expression was restricted to the islets (Fig 3B).
Figure 2. Comparison of Fto and Ftm expression in various tissue types.
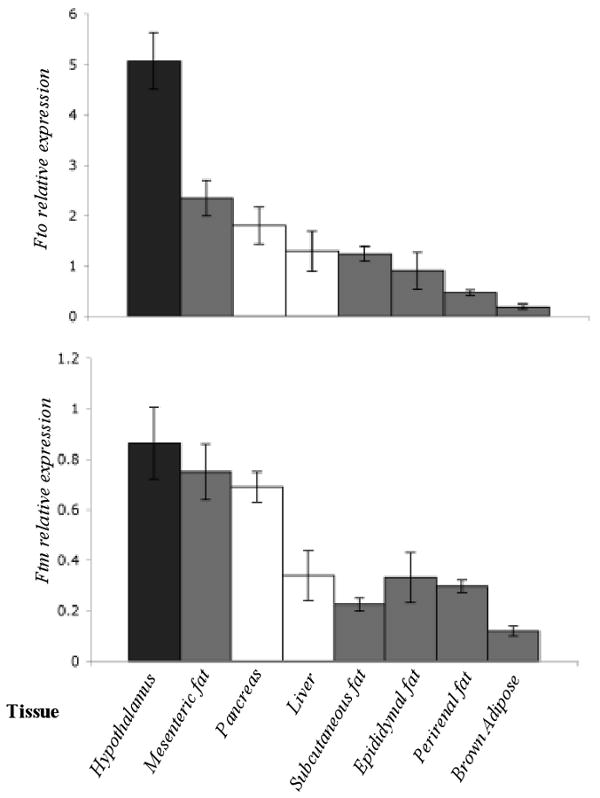
Fto and Ftm expression quantified by qPCR in the hypothalamus, mesenteric fat, pancreas, liver, subcutaneous fat, epididymal fat, perirenal fat and brown adipose tissue from 4-week +/+ C57BL/6J mice (N=5). Transcript levels were normalized to Gapdh.
Figure 3. Localization of Fto and Ftm transcripts in adult hypothalamus, pancreas and 13.5 dpc embryo.
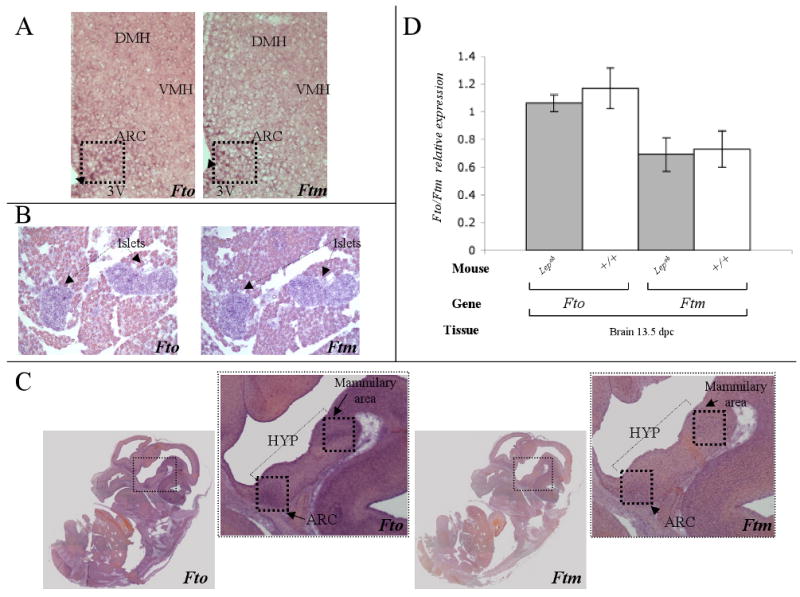
Fto and Ftm expression by in situ hybridization in (A) medial coronal hypothalamic and (B) sagittal pancreatic sections from 4-week old C57BL/6J males. Positive alkaline phosphatase staining of Fto and Ftm transcript is seen as a dark perinuclear ring. Fto is expressed in the arcuate nucleus (ARC), the dorsal medial hypothalamus (DMH), and the ventral medial hypothalamus (VMH). Ftm expression is restricted to the ARC in the hypothalamus and is expressed at a lower level than Fto. (C) Fto and Ftm in situ hybridization in medial sagittal sections of 13.5 dpc wild type embryos. Positive alkaline phosphatase staining of Fto and Ftm is seen as a dark perinuclear ring. Fto expression is present in the whole embryo, particularly in the brain and spinal cord. In the brain, Fto expression is enriched in the arcuate nucleus (ARC) and mamillary area. Ftm is expressed mainly in the brain and appears restricted to the ARC and mammilary area. (D) Fto and Ftm expression measured by Real Time PCR in whole brains of 13.5 dpc Lepob and +/+ mice. Transcript levels were normalized to Gapdh.
Effects of Lepob, Leprdb and Ay mutations on Fto and Ftm expression
Fto and Ftm expression were decreased by ∼2 to 3-fold (p<0.01) in the mesenteric fat and liver of 4-week old Ay and Lepob mice compared to the +/+ controls (Fig 4A, 4B). In subcutaneous, epididymal, renal and brown fat tissues, Fto and Ftm expression followed a similar trend. In pancreas, Fto and Ftm expression did not differ between lean and obese mice (Fig 4). By in situ hybridization, Fto and Ftm expression in sections of Lepob pancreata was comparable to that of +/+ mice (data not shown), and also limited to the islets. As anticipated, expression levels in Leprdb mice were comparable to those in Lepob (Fig. 5B)
Figure 4. Effects of obesity mutations in Fto and Ftm expression.
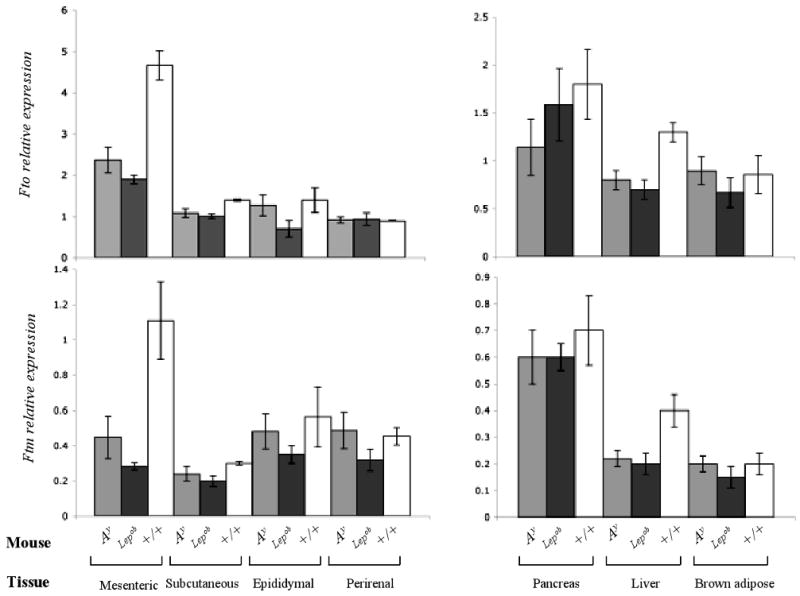
Fto and Ftm expression measured by qPCR in various tissues of Ay, Lepob and +/+ (C57BL/6J) mice. Transcript levels were normalized with Gapdh. N=5 each group.
Figure 5. Comparison of Fto and Ftm expression in various tissues and obesity models.
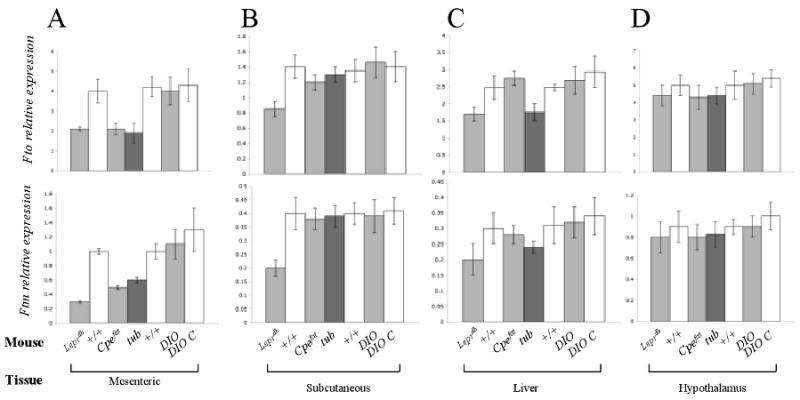
Fto and Ftm expression quantified by qPCR in (A) mesenteric fat, (B) subcutaneous fat, (C) liver and (D) hypothalamus of Leprdb, Cpefat, tub, Diet Induced Obese C57BL/6J (DIO), DIO C57BL/6J control (DIO C) and chow-fed +/+ (C57BL/6J) mice. Transcript levels were normalized with Gapdh. N=5 each group.
Effects of Cpefat and tub mutations, and DIO on Fto and Ftm expression
Fto and Ftm expression was assessed in mesenteric fat, liver subcutaneous fat, and hypothalamus of 4-week old Cpefat, tub and DIO mice. Expression of Fto and Ftm was reduced ∼2-fold in the mesenteric fat of Cpefat and tub mice. There was no statistically significant difference in the expression of either gene in any organ from DIO mice (Fig 5A). Fto expression was also reduced by 30% (p<0.03) in the liver of tub mice, and Ftm expression followed the same trend (Fig. 5C). There was no significant difference in Fto or Ftm expression in subcutaneous fat or liver of Cpefat, tub and DIO compared to lean mice.
Fto and Ftm expression in hypothalami and mesenteric fat of fed, fasted and 4°C ambient animals
In hypothalamus, Fto and Ftm expression did not differ among ad libitum fed Ay, Lepob, Leprdb, Cpefat, tub or DIO compared to fed control mice (Fig 5D, 6A). By in situ hybridization, Fto and Ftm expression in hypothalamic sections of Lepob mice was identical to that of +/+ mice (data not shown). To assess other possible regulatory functions of Fto /Ftm, their expression was measured in fasted animals and mice exposed to 4 °C for 30 minutes. Fto and Ftm expression levels was decreased in hypothalami of fasted Lepob mice compared to fed Lepob (Fto -20%, p<0.03; Ftm -40%, p<0.001) and fed +/+ mice (Fto -20%, p<0.04; Ftm -45%, p<0.01). Fasting had no effect on Fto or Ftm expression in mesenteric fat of Lepob mice (Fig 6B). Fasting was associated with a 2-fold (p<0.01) decrease in Fto expression in mesenteric fat of +/+ mice, but had no effect on Ftm.
Figure 6. Comparison of Fto and Ftm expression in fed/fasted mice.
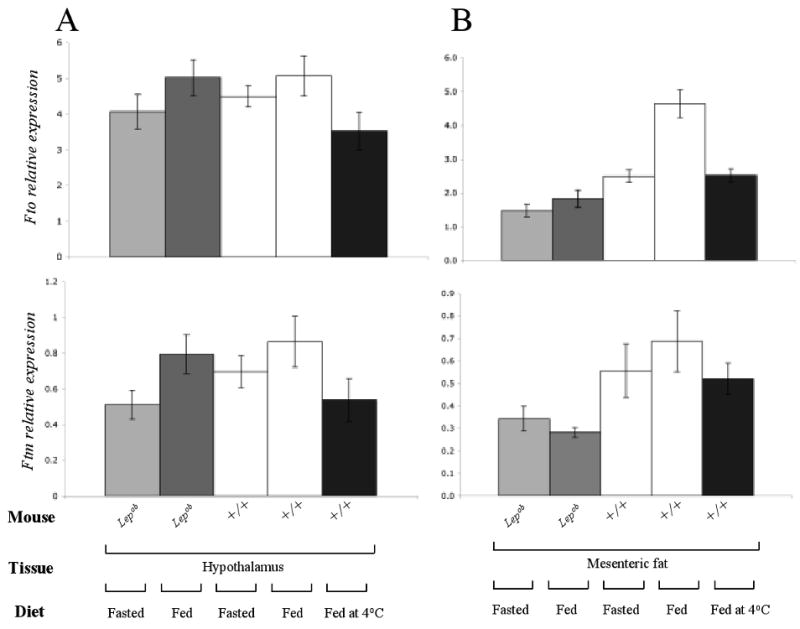
Fto and Ftm expression measured by qPCR in (A) hypothalamus and (B) mesenteric fat of fed/fasted Lepob, +/+ (C57BL/6J) mice and +/+ mice fed at 4°C for 30min. Transcript levels were normalized to Gapdh. N=5 each group.
Fto and Ftm expression decreased by 30% (p<0.01) and 40% (p<0.04), respectively, in hypothalami of +/+ mice exposed to 4°C for 30 minutes (Fig 6A). In the same mice, Fto expression was reduced 2-fold in mesenteric fat (p<0.001); there was a similar trend in Ftm expression (Fig 6A).
Comparison of Fto/Ftm expression in adipocytes and stromal vascular cells (SVC)
To determine whether Fto/Ftm are expressed in both adipocytes and SVC, cDNA specific to each cell fraction obtained from epididymal fat of +/+ and Leprdb mice (68) was used as template for qPRC analysis. Fto and Ftm were expressed in both adipocytes and SVC, though both genes were expressed at ∼36% (p=0.005) and ∼26% (p=0.01) higher levels in SVC, respectively (Fig. 7). The expression of both genes was decreased in both adipocytes (Fto, ∼60%, p=0.007; Ftm, ∼45%, p=0.01) and SVC (Fto, ∼67%, p<0.001; Ftm, ∼69%, p<0.001) obtained from Leprdb compared to +/+ mice (Fig. 7).
Figure 7. FTO and FTM are expressed in adipocytes and stromal vascular cells.
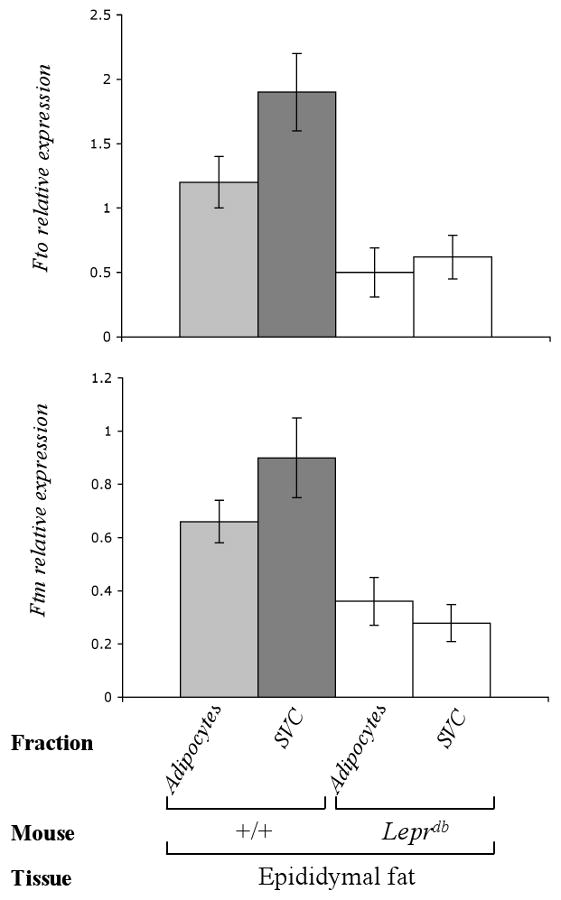
Fto/Ftm expression determined by qPCR in adipocytes and stromal vascular cells (SVC) from epididymal fat of +/+ (C57BL/6J) and Leprdb mice (68). N=2 each group
Effects of adiposity per se on Fto/Ftm expression
To assess the respective contributions to Fto/Ftm expression of 1.) obesity mutation(s), per se, and 2.) the degree of adiposity resulting from such mutations, we assessed body fat content by TD-NMR in Lepob, Leprdb, Cpefat and tub animals at 4-weeks of age, and obtained body composition data for 18-week old DIO and DIO control animals from The Jackson Laboratory (Maine, USA). Data grouped by tissue or mutation/DIO showed statistical significance (ANOVA; p<0.001). Significant tissue × mutation/DIO interactions were also present for both Fto and Ftm (ANOVA; p<0.001). We used absolute and fractional fat content to adjust (by ANCOVA) expression levels of Fto/Ftm in all tissues, by genotype. Levels of adiposity, per se, had no significant effect on these expression differences. Hence, expression differences are generally attributable to the respective obesity mutations or DIO.
Fto and Ftm expression in the mouse embryo
Fto and Ftm expression were also examined by in situ hybridization and qPCR in the mouse embryo. At 13.5 dpc, Fto was expressed throughout the whole embryo, especially in the brain and spinal cord (Fig 3C). In the midbrain, Fto expression was relatively high in the developing arcuate nucleus and mamillary area. Ftm was expressed at lower levels in fewer tissues than Fto. The Ftm expression pattern in the midbrain was comparable to that of Fto (Fig 3C). In whole brain, Ftm expression levels were ∼2-fold less than Fto (p<0.02) (Fig 3D).
The role of leptin during embryonic development is not clear. Leptin receptor isoforms are expressed in the mouse brain as early as 13.5 dpc and seem to induce differentiation of neuronal lineage cells (62). Fto and Ftm expression were assessed in Lepob embryos. The Lepob mutation was not associated with differences in Fto or Ftm expression in the whole brain of embryos at 13.5 dpc in comparison to +/+ controls (Fig 3D). Similarly, no differences in patterns of Fto or Ftm expression were observed between Lepob and +/+ embryos at 13.5 dpc by in situ hybridization (data not shown).
CUTL1 controls FTO and FTM expression
All FTO SNPs reported to be strongly associated with BMI (8, 14, 51, 52), and all 46 SNPs in linkage disequilibrium with those associated SNPs, were analyzed using MatInspector (matrix CDPCR3.01. Genomatix GmbH, Germany; http://www.genomatix.de) to identify canonical cis transcriptional regulatory elements containing these SNPs. rs17817449 (Matrix similarity 0.78) and rs8050136 (Matrix similarity 0.82) (Fig 1), were predicted to be located in CUTL1 binding sites (Fig. 8A). In chromatin immunoprecipitation (ChIP) of DNA from human fibroblasts using a CUTL1-specific antibody, a ∼90bp fragment that included rs8050136 was precipitated (Fig. 8B). Since the fibroblasts were heterozygous for rs8050136(A/C), it was possible to determine whether CUTL1 displayed a binding preference for either of these alleles. Only 20% of the rs8050136 fragments isolated by ChIP with the CUTL1 antibody carried the “C” allele (Fig 8C). When CUTL1 expression in the fibroblasts was reduced by 70% with siRNA, FTO expression was decreased by 90% and FTM by 65% (p<0.001) (Fig. 8D).
Figure 8. CUTL1 regulates the transcription of FTO and FTM.
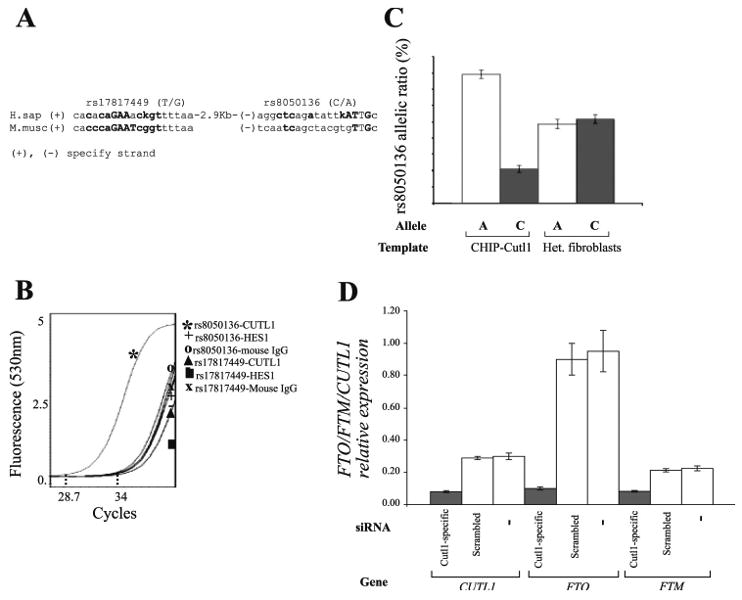
(A) rs17817449 and rs8050136 are located within CUTL1 recognition sequences as identified by MatInspector (Genomatix; http://www.genomatix.de). Complementary mouse sequence is aligned to the human sequence. Bases that fit the consensus are highlighted. (B) Lightcycler chromatogram showing the crossing point of PCR reactions containing DNA fragments isolated by ChIP with CUTL1, HES1 and mouse IgG antibodies. (C) Pyrosequencing using DNA extracts isolated by ChIP with the Cutl1 antibody. Genomic DNA isolated from fibroblasts heterozygous for rs8050136 (A/C) was used as control. (D) Expression analysis of CUTL1, FTO and FTM in human fibroblasts heterozygous for rs8050136 (A/C) where CUTL1 has been knocked down with siRNA.
Discussion
In all of the obesity mutations studied, Fto and Ftm expression was lower in the mesenteric fat than any other fat depot. The reasons for preferential effects (versus other adipose tissue depots) in the mesenteric adipose tissue, or for the higher levels of Fto and Ftm expression in SVC than adipocytes are not clear. In a recent study of human adipose tissue, FTO expression was reported to be about 2-fold higher in isolated adipocytes than whole subcutaneous adipose tissue, implying that in humans expression in adipocytes is greater than SVC (66). In that study, obesity increased expression in isolated adipocytes to a greater extent than in total adipose tissue. Further studies will have to be done to determine whether relative rates of expression in adipose tissue subfractions and effects of obesity on expression differ between mice and humans.
Downregulation of Fto, but not Ftm, in the mesenteric fat of fasted +/+ animals is effectively the only qualitative difference in expression pattern between the two genes. The decline in Fto expression in this depot with fasting (a response not seen in other fat depots) suggests that levels of FTO/FTM expression may be influenced by substrates coming at high concentrations from the small bowel.
In situ hybridization suggests that Ftm and Fto are co-expressed in a limited region of the arcuate nucleus. In the Allen Brain Atlas (http://www.brainatlas.org/aba/), Cutl1 spatial expression is comparable to that of Ftm in the arcuate nucleus of the adult mouse. In addition, the expression pattern in fasted animals is consistent with downregulation of Fto and Ftm expression observed in the hypothalamus of +/+ mice housed at 4°C in that cold exposure increases food consumption in mice (9). These findings are consistent with centrally-mediated effects on energy homeostasis. Such influence could be conveyed by developmental/structural effects of FTO/FTM and/or participation -as suggested by responses to environmental manipulation- in intercurrent metabolic/behavioral homeostasis. The salience of effects in the genetic models with direct interruptions of the leptin axis (Lepob, Leprdb) are intriguing, and could point to specific neurophysiological roles for Fto and Ftm in canonical neuroregulation of energy metabolism. The site-related differences in adipose tissue expression, could reflect cell-autonomous differences in Fto/Ftm expression and/or neurally-mediated effects on these depots. The apparent role of FTO/FTM in cell cycle and developmental aspects of the hypothalamus might suggest that they would be unlikely to respond to intercurrent metabolic or environmental changes. However, there is ample precedent for just such a dual role (in brain development and metabolic homeostasis) by hormone leptin (5).
Following completion of the present study, Gerken et al (15) reported that the FTO protein has sequence similarity to Fe(II)- and 2-oxoglutarate –dependent oxygenases, localizes to the nucleus, and demethylates single-stranded DNA in vitro in the presence of Fe(II) and ascorbate, suggesting a possible role for FTO in regulation of gene transcription or DNA damage repair. They reported a 60% decrease of Fto expression by qPCR in laser-dissected murine arcuate nuclei of fasted animals, but no alteration in ventromedial or paraventricular nuclei of these same mice. In our study, we also observed a trend towards reduced Fto expression levels in whole hypothalami of fasted +/+ mice, and found a statistically significant decrease (∼20%) in fasted Lepob compared to fed Lepob mice (Fig. 6A). Our data are consistent with those of Gerken et al (15), and suggest that leptin does not mediate these declines of Fto expression in fasted animals.
Shh expression is reduced in the limb buds of Ftm-/- mouse embryos (65). In chick limb buds, Shh overexpression causes ectopic expression of Cux2, a Cutl1 ortholog (58). Thus, a positive feedback loop may exist between Cutl1, Ftm and SHH signaling. Misregulation of SHH signaling and cilia dysfunction are implicated in obesity, retinal degeneration, right-left asymmetry, renal dysplasia and polydactyly that characterize the Bardet-Biedl syndrome (10, 57, 59). Ftm is a basal body protein of cilia that affects SHH signaling (65), suggesting that Ftm may contribute to aspects of hypothalamic development. Reminiscent of aspects of the Bardet-Biedl syndrome, humans with mutations in RPGRIP, the FTM homologue, develop retinal dysplasia (43, 57). Additionally, Cutl1-/- and Fused toes homozygous mutants display left-right asymmetry.
There is striking similarity of gene structure and order of Fto, Ftm, Fts and the Irx genes between human and mouse (Fig 1). Moreover, the large Fto intronic regions (up to ∼177.5 Kb) show extended sequence conservation with their human counterparts (Genomic Sequence Alignment function; Ensembl). SNP rs8050136, and the CUTL1 binding site in which it is located, are part of a highly conserved 1.2 Kb intronic region (58% identity with the mouse). CUTL1 belongs to the CDP/Cut family of homeoproteins (40). It consists of a Cut homeodomain and 3 ‘Cut repeat’ DNA-binding domains (18). Members of the CDP/Cut family have the capacity to bind to a wide range of DNA sequences (1, 3, 7, 18, 64). In the present study, CUTL1 interacted only with the predicted binding sequence “aggctcagatatt(g/t)ATTGc” (rs8050136) and not with the predicted binding sequence “cacacaGAAac(g/t)gttttaa” (rs17817449) (Fig 8A). Moreover, CUTL1 preferentially bound to DNA fragments carrying the ‘C’ allele of rs8050136. Using data from the Tubingen Family Study, Tschritter et al (61) identified a dose-dependent increase of ∼5 Kg in body mass.
Cutl1 was originally cloned in Drosophila and shown to be a downstream effector of Notch (24, 34, 38, 42). Lack of functional Cutl1 is embryonic lethal, while flies with viable mutations display developmental defects in various organs including limbs, Malpighian tubules and external sensory organs (4, 23, 30). Cutl1 was originally identified in vertebrates for its CCAAT displacement activity and was later linked by homology to its Drosophila paralog (41). In the mouse, disruption of Cutl1 results in generalized somatic growth retardation (12, 31, 53), while Cutl1 overexpression leads to multiorgan cellular hyperplasia and organomegaly (26). CUTL1 plays an apparent role in cell cycle progression, specifically as an accelerator of entry into S phase (50).
CUTL1 acts as a transcriptional repressor by displacing activators (54) and/or by recruitment of histone deacetylase 1 (33), and has been proposed to inhibit gene expression in terminally differentiated cells (13). However, CUTL1 has also been implicated as a transcriptional activator. This activity depends upon proteolytic cleavage, resulting in a truncated isoform, p110 (including the C-terminal ”Cut repeats“ 2,3 and the homeodomain), that upregulates DNA pol a (60). p110 and the DNA pol a promoter interact during the G1/S phase transition (50, 60). In the present study, the ChIP assay was performed using an antibody that recognizes both CUTL1 and p110 (60). The siRNA knock-down experiment indicates that CUTL1 (or p110) is needed for FTO and FTM transcriptional activation. The finding that FTO and FTM are co-regulated by CUTL1 is consistent with the expression data in mouse organs. The discovery of the genetic interaction between CUTL1 and FTO/FTM at rs8050136 does not exclude the possibility that regulation by CUTL1 may involve other CUTL1 binding sites. In the implicated (by LD) ∼47Kb region, MatInspector (Genomatix; http://www.genomatix.de) identified 12 CUTL1 binding sites (matrix CLOX/CDPCR3.01; Matrix similarity >0.8) all present in the first intron of FTO. Except rs8050136, only SNP rs7202296 (∼6 Kb downstream of rs8050136) is located in a putative CUTL1 binding site.
Perspectives and Significance
The recent availability of very high density molecular maps of intrinsic single nucleotide pair variation within the human genome has enabled a new means of looking for genes that account for quantitative (adiposity) or qualitative (diabetes) human phenotypic variation. In such genome-wide scans (GWAS), no prior information regarding the type of gene or its physical location is required. Statistical signals correlating phenotype with SNPs are used to implicate genetic regions and constituent genes. These approaches, while inherently more sensitive than older parametric linkage techniques, can (and do) generate large numbers of authentic “candidates” whose functional relevance with regard to the dependent phenotype is largely or entirely unknown. FTO (FTM) are good examples. One or both of these genes are clearly implicated (statistically) in control of human adiposity (BMI), but the molecular physiology of such effects is unknown. FTO is apparently an Fe(II)- and 2-oxoglutarate –dependent oxygenase, and FTM a ciliary basal body component. The former might operate as a DNA demethylase, the latter in what is now appreciated - by virtue of the obesity phenotype in the Bardet-Biedl and Alstrom syndromes - as an important pathway in human energy homeostasis. To get an idea of how these newly implicated genes might operate in energy homeostasis, we examined their organ distributions (ubiquitous); their changes in expression in the context of classical monogenic and dietary obesities (reduced), and in response to food restriction and cold exposure (decreased); and the distribution of their transcripts within the arcuate of the hypothalamus. While these findings are consistent with a role for these genes in canonical molecular pathways related to energy homeostatsis, the specific mechanisms/pathways by which one or both genes influence adiposity are not revealed by our experiments. The study of mice with conditional, organ-specific knockouts/overexpression of these genes (alone and together) will be needed. As will studies of the expression of FTO/FTM in relevant tissues obtained from humans with 0,1, or 2 of the risk alleles for rs8050136. More interesting at this point is the possibility that both genes are being regulated by a single transcription factor (CUTL1) via a single regulatory site in the first intron of FTO. While our paper provides circumstantial evidence in this regard, more definitive analysis will require the study of mice with under-/over-active alleles of Cutl, and, hopefully, the identification of humans with aberrant alleles of CUTL1. Whereas the mouse has been heavily relied upon to identify candidate genes for phenotypes such as obesity and diabetes, in the era of the GWAS, their role will now be expanded to the vetting of mechanisms for genes discovered in high resolution “sweeps” of the human genome.
Supplementary Material
Acknowledgments
The authors thank Kourosh Ahmadi (King's College, London) for assistance with bioinformatics and Patricia Lanzano (Naomi Berrie Diabetes Center, Columbia University) for cell culture assistance. The authors also thank Yiying Zhang for generously providing cDNA from epididymal depot cellular fractions.
GRANTS: This work was funded by RO1 DK52431-15, P30 DK6687-26 and an ADA mentored fellowship award. Work was also supported by the European Commission for the MolPAGE Consortium (LSHG-CT-2004-512066). Andrew Hattersley is a Wellcome Trust Research Leave Fellow.
References
- 1.Andres V, Chiara MD, Mahdavi V. A new bipartite DNA-binding domain: cooperative interaction between the cut repeat and homeo domain of the cut homeo proteins. Genes Dev. 1994;8:245–57. doi: 10.1101/gad.8.2.245. [DOI] [PubMed] [Google Scholar]
- 2.Anselme I, Laclef C, Lanaud M, Ruther U, Schneider-Maunoury S. Defects in brain patterning and head morphogenesis in the mouse mutant Fused toes. Dev Biol. 2007;304:208–20. doi: 10.1016/j.ydbio.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 3.Aufiero B, Neufeld EJ, Orkin SH. Sequence-specific DNA binding of individual cut repeats of the human CCAAT displacement/cut homeodomain protein. Proc Natl Acad Sci U S A. 1994;91:7757–61. doi: 10.1073/pnas.91.16.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodmer R, Barbel S, Sheperd S, Jack JW, Jan LY, Jan YN. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell. 1987;51:293–307. doi: 10.1016/0092-8674(87)90156-5. [DOI] [PubMed] [Google Scholar]
- 5.Bouret SG, Draper SJ, Simerly RB. Trophic action of leptin on hypothalamic neurons that regulate feeding. Science. 2004;304:108–10. doi: 10.1126/science.1095004. [DOI] [PubMed] [Google Scholar]
- 6.Braissant O, Wahli W. Simplified in situ hybridization protocol using non-radioactivity labeled probes to detect abundant and rare mRNAs on tissue sections. Biochemica. 1998;1:10–16. [Google Scholar]
- 7.Catt D, Luo W, Skalnik DG. DNA-binding properties of CCAAT displacement protein cut repeats. Cell Mol Biol (Noisy-le-grand) 1999;45:1149–60. [PubMed] [Google Scholar]
- 8.Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, Delplanque J, Vaillant E, Pattou F, Ruiz J, Weill J, Levy-Marchal C, Horber F, Potoczna N, Hercberg S, Le Stunff C, Bougneres P, Kovacs P, Marre M, Balkau B, Cauchi S, Chevre JC, Froguel P. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007 May 13;39:724–6. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 9.Coleman DL. Thermogenesis in diabetes-obesity syndromes in mutant mice. Diabetologia. 1982;22:205–11. doi: 10.1007/BF00283754. [DOI] [PubMed] [Google Scholar]
- 10.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–66. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davenport JR, Watts AJ, Roper VC, Croyle MJ, van Groen T, Wyss JM, Nagy TR, Kesterson RA, Yoder BK. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr Biol. 2007;17:1586–94. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis T, Gambardella L, Horcher M, Tschanz S, Capol J, Bertram P, Jochum W, Barrandon Y, Busslinger M. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 2001;15(2):307–19. doi: 10.1101/gad.200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fei X, Qin Z, Liang Z. Contribution of CDP/Cux, a transcription factor, to cell cycle progression. Acta Biochim Biophys Sin (Shanghai) 2007;39:923–30. doi: 10.1111/j.1745-7270.2007.00366.x. [DOI] [PubMed] [Google Scholar]
- 14.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, Galvanovskis J, Rorsman P, Robins P, Prieur X, Coll AP, Ma M, Jovanovic Z, Farooqi IS, Sedgwick B, Barroso I, Lindahl T, Ponting CP, Ashcroft FM, O'Rahilly S, Schofield CJ. The Obesity-Associated FTO Gene Encodes a 2-Oxoglutarate Dependent Nucleic Acid Demethylase. [Online] Science. 2007 doi: 10.1126/science.1151710. http://www.sciencemag.org/cgi/content/abstract/1151710v1. [DOI] [PMC free article] [PubMed]
- 16.Gotz K, Briscoe J, Ruther U. Homozygous Ft embryos are affected in floor plate maintenance and ventral neural tube patterning. Dev Dyn. 2005;233:623–30. doi: 10.1002/dvdy.20354. [DOI] [PubMed] [Google Scholar]
- 17.Grotewold L, Ruther U. The Fused toes (Ft) mouse mutation causes anteroposterior and dorsoventral polydactyly. Dev Biol. 2002;251:129–41. doi: 10.1006/dbio.2002.0817. [DOI] [PubMed] [Google Scholar]
- 18.Harada R, Berube G, Tamplin OJ, Denis-Larose C, Nepveu A. DNA-binding specificity of the cut repeats from the human cut-like protein. Mol Cell Biol. 1995;15:129–40. doi: 10.1128/mcb.15.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, Colditz G, Hinney A, Hebebrand J, Koberwitz K, Zhu X, Cooper R, Ardlie K, Lyon H, Hirschhorn JN, Laird NM, Lenburg ME, Lange C, Christman MF. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–83. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 20.Heymer J, Kuehn M, Ruther U. The expression pattern of nodal and lefty in the mouse mutant Ft suggests a function in the establishment of handedness. Mech Dev. 1997;66:5–11. doi: 10.1016/s0925-4773(97)00084-1. [DOI] [PubMed] [Google Scholar]
- 21.Hinney A, Nguyen TT, Scherag A, Friedel S, Brönner G, Müller TD, Grallert H, Illig T, Wichmann HE, Rief W, Schäfer H, Hebebrand J. Genome Wide Association (GWA) Study for Early Onset Extreme Obesity Supports the Role of Fat Mass and Obesity Associated Gene (FTO) Variants. PLoS ONE. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong DH, Yue G, Adamian M, Li T. Retinitis pigmentosa GTPase regulator (RPGRr)-interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium. J Biol Chem. 2001;276:12091–9. doi: 10.1074/jbc.M009351200. [DOI] [PubMed] [Google Scholar]
- 23.Jack J, DeLotto Y. Effect of wing scalloping mutations on cut expression and sense organ differentiation in the Drosophila wing margin. Genetics. 1992;131:353–63. doi: 10.1093/genetics/131.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson TK, Judd BH. Analysis of the Cut Locus of DROSOPHILA MELANOGASTER. Genetics. 1979;92:485–502. doi: 10.1093/genetics/92.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar J, Sunkishala RR, Karthikeyan G, Sengupta S. The common genetic variant upstream of INSIG2 gene is not associated with obesity in Indian population. Clin Genet. 2007;7:415–8. doi: 10.1111/j.1399-0004.2007.00795.x. [DOI] [PubMed] [Google Scholar]
- 26.Ledford AW, Brantley JG, Kemeny G, Foreman TL, Quaggin SE, Igarashi P, Oberhaus SM, Rodova M, Calvet JP, Vanden Heuvel GB. Deregulated expression of the homeobox gene Cux-1 in transgenic mice results in downregulation of p27(kip1) expression during nephrogenesis, glomerular abnormalities, and multiorgan hyperplasia. Dev Biol. 2002;245:157–71. doi: 10.1006/dbio.2002.0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Wu Y, Loos RJ, Hu FB, Liu Y, Wang J, Yu Z, Lin X. Variants in the fat mass- and obesity-associated (FTO) gene are not associated with obesity in a Chinese Han population. Diabetes. 2008;57:264–8. doi: 10.2337/db07-1130. [DOI] [PubMed] [Google Scholar]
- 28.Leibel RL, Chua SC, Rosenbaum M. Obesity: : The Molecular Physiology of Weight Regulation. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabol Basis of Inherited Disease. VIII. 2001. pp. 3965–4028. [Google Scholar]
- 29.Leibel RL, Chung WK, Chua SC., Jr The molecular genetics of rodent single gene obesities. J Biol Chem. 1997;272:31937–40. doi: 10.1074/jbc.272.51.31937. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, Jack J. Regulatory interactions and role in cell type specification of the Malpighian tubules by the cut, Kruppel, and caudal genes of Drosophila. Dev Biol. 1992;150:133–43. doi: 10.1016/0012-1606(92)90013-7. [DOI] [PubMed] [Google Scholar]
- 31.Luong MX, van der Meijden CM, Xing D, Hesselton R, Monuki ES, Jones SN, Lian JB, Stein JL, Stein GS, Neufeld EJ, van Wijnen AJ. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol Cell Biol. 2002;22:1424–37. doi: 10.1128/mcb.22.5.1424-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyon HN, Florez JC, Bersaglieri T, Saxena R, Winckler W, Almgren P, Lindblad U, Tuomi T, Gaudet D, Zhu X, Cooper R, Ardlie KG, Daly MJ, Altshuler D, Groop L, Hirschhorn JN. Common variants in the ENPP1 gene are not reproducibly associated with diabetes or obesity. Diabetes. 2006;55:3180–4. doi: 10.2337/db06-0407. [DOI] [PubMed] [Google Scholar]
- 33.Mailly F, Berube G, Harada R, Mao PL, Phillips S, Nepveu A. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol Cell Biol. 1996;16:5346–57. doi: 10.1128/mcb.16.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majumdar A, Nagaraj R, Banerjee U. strawberry notch encodes a conserved nuclear protein that functions downstream of Notch and regulates gene expression along the developing wing margin of Drosophila. Genes Dev. 1997;11:1341–53. doi: 10.1101/gad.11.10.1341. [DOI] [PubMed] [Google Scholar]
- 35.Matsuoka N, Patki A, Tiwari HK, Allison DB, Johnson SB, Gregersen PK, Leibel RL, Chung WK. Association of K121Q polymorphism in ENPP1 (PC-1) with BMI in Caucasian and African-American adults. Int J Obes (Lond) 2006;30:233–7. doi: 10.1038/sj.ijo.0803132. [DOI] [PubMed] [Google Scholar]
- 36.Mavlyutov TA, Zhao H, Ferreira PA. Species-specific subcellular localization of RPGR and RPGRIP isoforms: implications for the phenotypic variability of congenital retinopathies among species. Hum Mol Genet. 2002;11:1899–907. doi: 10.1093/hmg/11.16.1899. [DOI] [PubMed] [Google Scholar]
- 37.Meyre D, Bouatia-Naji N, Tounian A, Samson C, Lecoeur C, Vatin V, Ghoussaini M, Wachter C, Hercberg S, Charpentier G, Patsch W, Pattou F, Charles MA, Tounian P, Clement K, Jouret B, Weill J, Maddux BA, Goldfine ID, Walley A, Boutin P, Dina C, Froguel P. Variants of ENPP1 are associated with childhood and adult obesity and increase the risk of glucose intolerance and type 2 diabetes. Nat Genet. 2005;37:863–7. doi: 10.1038/ng1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124:1485–95. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- 39.Mykytyn K, Sheffield VC. Establishing a connection between cilia and Bardet-Biedl Syndrome. Trends Mol Med. 2004;10:106–9. doi: 10.1016/j.molmed.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- 41.Neufeld EJ, Skalnik DG, Lievens PM, Orkin SH. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet. 1992:50–5. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 42.Neumann CJ, Cohen SM. A hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wing. Development. 1996;122:3477–85. doi: 10.1242/dev.122.11.3477. [DOI] [PubMed] [Google Scholar]
- 43.Nishimura DY, Fath M, Mullins RF, Searby C, Andrews M, Davis R, Andorf JL, Mykytyn K, Swiderski RE, Yang B, Carmi R, Stone EM, Sheffield VC. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc Natl Acad Sci U S A. 2004;101:16588–93. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohashi J, Naka I, Kimura R, Natsuhara K, Yamauchi T, Furusawa T, Nakazawa M, Ataka Y, Patarapotikul J, Nuchnoi P, Tokunaga K, Ishida T, Inaoka T, Matsumura Y, Ohtsuka R. FTO polymorphisms in oceanic populations. J Hum Genet. 2007;52:1031–5. doi: 10.1007/s10038-007-0198-2. [DOI] [PubMed] [Google Scholar]
- 45.Pawlyk BS, Smith AJ, Buch PK, Adamian M, Hong DH, Sandberg MA, Ali RR, Li T. Gene replacement therapy rescues photoreceptor degeneration in a murine model of Leber congenital amaurosis lacking RPGRIP. Invest Ophthalmol Vis Sci. 2005;46:3039–45. doi: 10.1167/iovs.05-0371. [DOI] [PubMed] [Google Scholar]
- 46.Peeters A, Beckers S, Verrijken A, Roevens P, Peeters P, Van Gaal L, Van Hul W. Variants in the FTO gene are associated with common obesity in the Belgian population. [Online] Mol Genet Metab. 2007 doi: 10.1016/j.ymgme.2007.10.011. http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WNG-4R8PNV3-1&_user=18704&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000002018&_version=1&_urlVersion=0&_userid=18704&md5=2fbbdc150f8ed8246729353f74a24581. [DOI] [PubMed]
- 47.Peters T, Ausmeier K, Dildrop R, Ruther U. The mouse Fused toes (Ft) mutation is the result of a 1.6-Mb deletion including the entire Iroquois B gene cluster. Mamm Genome. 2002;13:186–8. doi: 10.1007/s00335-001-2142-7. [DOI] [PubMed] [Google Scholar]
- 48.Peters T, Ausmeier K, Ruther U. Cloning of Fatso (Fto), a novel gene deleted by the Fused toes (Ft) mouse mutation. Mamm Genome. 1999;10:983–6. doi: 10.1007/s003359901144. [DOI] [PubMed] [Google Scholar]
- 49.Raman RP. Obesity and health risks. J Am Coll Nutr. 2002;21:134S–139S. doi: 10.1080/07315724.2002.10719210. [DOI] [PubMed] [Google Scholar]
- 50.Sansregret L, Goulet B, Harada R, Wilson B, Leduy L, Bertoglio J, Nepveu A. The p110 isoform of the CDP/Cux transcription factor accelerates entry into S phase. Mol Cell Biol. 2006;26:2441–55. doi: 10.1128/MCB.26.6.2441-2455.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR. Genome-Wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinclair AM, Lee JA, Goldstein A, Xing D, Liu S, Ju R, Tucker PW, Neufeld EJ, Scheuermann RH. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood. 2001;98:3658–67. doi: 10.1182/blood.v98.13.3658. [DOI] [PubMed] [Google Scholar]
- 54.Skalnik DG, Strauss EC, Orkin SH. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J Biol Chem. 1991;266:16736–44. [PubMed] [Google Scholar]
- 55.Smith AJ, Cooper JA, Li LK, Humphries SE. INSIG2 gene polymorphism is not associated with obesity in Caucasian, Afro-Caribbean and Indian subjects. [Online] Int J Obes (Lond) 2007 doi: 10.1038/sj.ijo.0803645. http://www.nature.com/ijo/journal/v31/n11/full/0803645a.html. [DOI] [PubMed]
- 56.Stratakis CA, Lafferty A, Taymans SE, Gafni RI, Meck JM, Blancato J. Anisomastia associated with interstitial duplication of chromosome 16, mental retardation, obesity, dysmorphic facies, and digital anomalies: molecular mapping of a new syndrome by fluorescent in situ hybridization and microsatellites to 16q13 (D16S419-D16S503) J Clin Endocrinol Metab. 2000;85:3396–401. doi: 10.1210/jcem.85.9.6776. [DOI] [PubMed] [Google Scholar]
- 57.Takada T, Iida K, Sasaki H, Taira M, Kimura H. Expression of ADP-ribosylation factor (ARF)-like protein 6 during mouse embryonic development. Int J Dev Biol. 2005;49:891–4. doi: 10.1387/ijdb.052031tt. [DOI] [PubMed] [Google Scholar]
- 58.Tavares AT, Tsukui T, Izpisua Belmonte JC. Evidence that members of the Cut/Cux/CDP family may be involved in AER positioning and polarizing activity during chick limb development. Development. 2000;127:5133–44. doi: 10.1242/dev.127.23.5133. [DOI] [PubMed] [Google Scholar]
- 59.Tobin JL, Beales PL. Related ArticlesLinksBardet-Biedl syndrome: beyond the cilium. Pediatr Nephrol. 2007;22:926–36. doi: 10.1007/s00467-007-0435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Truscott M, Raynal L, Premdas P, Goulet B, Leduy L, Berube G, Nepveu A. CDP/Cux stimulates transcription from the DNA polymerase alpha gene promoter. Mol Cell Biol. 2003;23:3013–28. doi: 10.1128/MCB.23.8.3013-3028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tschritter O, Preissl H, Yokoyama Y, Machicao F, Haring HU, Fritsche A. Variation in the FTO gene locus is associated with cerebrocortical insulin resistance in humans. [Online] Diabetologia. 2007 doi: 10.1007/s00125-007-0839-1. http://www.nature.com/ijo/journal/v31/n11/full/0803645a.html. [DOI] [PubMed]
- 62.Udagawa J, Hatta T, Hashimoto R, Otani H. Related ArticlesLinksRoles of leptin in prenatal and perinatal brain development. Congenit Anom (Kyoto) 2007;47:77–83. doi: 10.1111/j.1741-4520.2007.00150.x. [DOI] [PubMed] [Google Scholar]
- 63.van den Ent FM, van Wijnen AJ, Lian JB, Stein JL, Stein GS. Cell cycle controlled histone H1, H3, and H4 genes share unusual arrangements of recognition motifs for HiNF-D supporting a coordinate promoter binding mechanism. J Cell Physiol. 1994;159:515–30. doi: 10.1002/jcp.1041590316. [DOI] [PubMed] [Google Scholar]
- 64.van der Hoeven F, Schimmang T, Volkmann A, Mattei MG, Kyewski B, Ruther U. Programmed cell death is affected in the novel mouse mutant Fused toes (Ft) Development. 1994;120:2601–7. doi: 10.1242/dev.120.9.2601. [DOI] [PubMed] [Google Scholar]
- 65.Vierkotten J, Dildrop R, Peters T, Wang B, Ruther U. Ftm is a novel basal body protein of cilia involved in Shh signalling. Development. 2007;134:2569–77. doi: 10.1242/dev.003715. [DOI] [PubMed] [Google Scholar]
- 66.Wåhlén K, Sjölin E, Hoffstedt J. The common rs9939609 gene variant of the fat mass and obesity associated gene (FTO) is related to fat cell lipolysis. [Online] J Lipid Res. 2007 doi: 10.1194/jlr.M700448-JLR200. http://www.jlr.org/cgi/reprint/M700448-JLR200v1. [DOI] [PubMed]
- 67.Weedon MN, Shields B, Hitman G, Walker M, McCarthy MI, Hattersley AT, Frayling TM. No evidence of association of ENPP1 variants with type 2 diabetes or obesity in a study of 8,089 U.K. Caucasians. Diabetes. 2006;55:3175–9. doi: 10.2337/db06-0410. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, Guo KY, Diaz PA, Heo M, Leibel RL. Determinants of leptin gene expression in fat depots of lean mice. Am J Physiol Regul Integr Comp Physiol. 2002;282:R226–34. doi: 10.1152/ajpregu.00392.2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.