Abstract
Hakai, an E3 ubiquitin ligase for the E-cadherin complex, plays a crucial role in lowering cell-cell contacts in epithelial cells, a hallmark feature of tumor progression. Recently, Hakai was also found to interact with PSF (PTB-associated splicing factor). While PSF can function as a DNA-binding protein with a tumor suppressive function, its association with Hakai promotes PSF’s RNA-binding ability and post-transcriptional influence on target mRNAs. Hakai overexpression enhanced the binding of PSF to mRNAs encoding cancer-related proteins, while knockdown of Hakai reduced the RNA-binding ability of PSF. Furthermore, the knockdown of PSF suppressed Hakai-induced cell proliferation. Thus, Hakai can affect the oncogenic phenotype both by altering E-cadherin-based intercellular adhesions and by increasing PSF’s ability to bind RNAs that promote cancer-related gene expression.
Keywords: E-cadherin, PSF, PTB-associated splicing factor, proliferation, tumor progression
Role of Hakai in cell proliferation and tumor progression
Hakai was originally identified as a RING finger-type E3 ubiquitin-ligase for the E-cadherin complex, a major component of adherens junctions. Hakai binds to the cytoplasmic domain of E-cadherin and mediates its ubiquitination, endocytosis and lysosomal degradation. In turn, the loss of E-cadherin leads to the disruption of epithelial cell-cell adhesions, a key event in epithelial transformation,1–6 and facilitates invasion and metastasis. The cytoplasmic domain of E-cadherin is connected to the cytoskeleton through proteins such as β-catenin and α-catenin, which regulate gene expression and participate in signaling pathways that modualte cell proliferation, differentiation, and survival.7,8 After activation of several tyrosine kinases, such as the receptors for EGF or HGF, the E-cadherin complex is phosphorylated at tyrosine residues. Phosphorylation of E-cadherin facilitates its recognition by Hakai, followed by its proteolysis and clearance from cell-cell contacts.
The occurrence of the excessive internalization and degradation of E-cadherin upon Hakai overexpression raises the possibility that Hakai could enhance tumor progression and metastasis.1,9 Several lines of evidence in our recent report also support a role of Hakai in tumorigenesis10: 1) Overexpression of Hakai promotes proliferation of various cultured cell lines; 2) Hakai is upregulated in hyperproliferative human endometrium and lymph nodes; 3), Hakai induces anchorage-independent cell growth, and 4) Hakai expression is elevated in human colon and gastric adenocarcinomas. Collectively, these data suggest that Hakai may be involved in tumorigenesis by disrupting intercellular adhesions and by promoting cell proliferation.
Hakai interacts with PSF and promotes PSF RNA-binding function
In search of downstream effectors of Hakai that function independently of binding to E-cadherin, we recently identified a novel Hakai-interacting protein, PSF (polypyrimidine tract-binding protein-associated splicing factor).10 PSF is a nuclear protein implicated in transcription, DNA binding, unwinding, and repair, as well as pre-mRNA splicing and RNA editing.11–19 PSF contains an N-terminal proline- and glutamine-rich domain, through which it interacts with Hakai, two RNA recognition motifs (RRMs), and a C-terminal region that contains two nuclear localization signals, both of which are required for its nuclear localization.20
PSF co-localizes with Hakai in the nucleus, raising the possibility that Hakai could play a role in the nucleus through its association with PSF. Previously, few specific mRNAs had been reported to be PSF targets. In our recent study, we demonstrated that Hakai overexpression influences the RNA-binding ability of PSF to specific target transcripts. By using ribonucleoprotein immunoprecipitation (RNP IP) assays using anti-PSF and control IgG antibodies, populations of PSF-associated mRNAs were extracted from the IP samples and reverse transcribed; the resulting probes were used to hybridize human cDNA arrays. This analysis revealed that overexpression of Hakai leads to the increased association of PSF with a significant number of transcripts.10
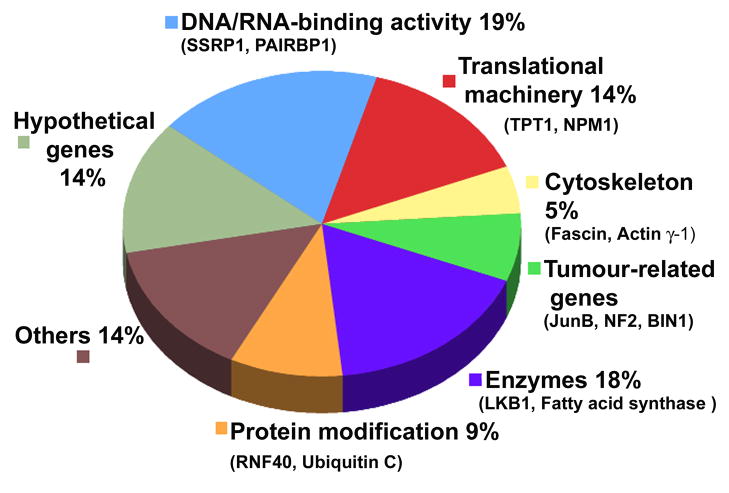
Several PSF-associated transcripts encode proteins implicated in protein translation and modification, tumorigenesis, cell adhesion, and cytoskeleton dynamics (Figure 1). The influence of Hakai in regulating the association of PSF with target mRNAs was also examined by analyzing Hakai knockdown cells. We found that silencing Hakai led to a reduction in PSF binding to target mRNAs, further supporting the notion that Hakai promotes the RNA-binding ability of PSF. Interestingly, we observed that PSF in Hakai-overexpressing cells migrated more slowly in polyacrylamide gels than it did in parental cells, suggesting that PSF was posttranslationally modified by Hakai. However, Hakai did not appear to induce ubiquitination of PSF, suggesting that PSF is not a substrate of this E3 enzyme. It is known that Mnk (the mitogen-activated protein kinase signal-integrating kinase) can phosphorylate PSF, an event that favours the binding of PSF to the TNF-α mRNA.21. NPM/ALK (tyrosine kinase nucleophosmin/anaplastic lymphoma kinase) also phosphorylates PSF, increasing its RNA-binding ability and inhibiting the function of PSF as a transcriptional repressor.22 Additionally, phosphorylation of PSF by BRK (breast tumor kinase) lowers PSF binding to bind RNA and induces the S-phase cell cycle arrest.23 It is possible that Hakai overexpression affects the activity of proteins that induce a modification on PSF, which in turn may modulate PSF’s interaction with target mRNAs and alter their post-transcriptional fate. According to this hypothesis, increases in Hakai abundance or in the activity of post-translational modulators of PSF can alter PSF’s properties from DNA-binding to RNA-binding, and consequently modulate cellular processes such as cell cycle arrest, proliferation or apoptosis.10, 21–23
Figure 1. Functional distribution of subsets of mRNAs bound to PSF in Hakai-overexpressing epithelial MDCK cells.
No transcripts were significantly enriched in association with PSF in normal epithelial MDCK cells. Examples of transcripts enriched in each category are shown.
Perspective: implication of PSF in proliferation and oncogenesis
In order to understand the physiological significance of the Hakai-PSF interaction, we focused on the process of cell proliferation. Our data suggested that Hakai may regulate cell proliferation by modulating PSF activity. In Hakai-overexpressing MDCK (Madin-Darby canine kidney) cells, stable knockdown of PSF partially suppressed the proliferative influence of Hakai overexpression. Moreover, expression of a Hakai mutant lacking the RING finger domain suppressed cell proliferation, suggesting that the E3 ubiquitin-ligase activity of Hakai is necessary for promoting cell division. This Hakai mutant was able to bind PSF, indicating that Hakai’s interaction with PSF alone did not promote cell proliferation. The physical and functional interactions between PSF and the E3 ubiquitin-ligase activity of Hakai await further analysis.
Several lines of evidence support a role for PSF in proliferation and oncogenesis. First, PSF promotes the translation of the cancer-related protein Myc in vitro and in vivo by binding to the Myc mRNA internal ribosome entry site (IRES) and increasing its activity. Second, PSF contains a DNA-binding domain that co-ordinately represses multiple oncogenic genes in human cell lines, suggesting a role for PSF in tumor suppression; however, binding of the retrotransposon VL30 RNA to PSF was followed by PSF’s disengagement from the repressed genes, to their increased transcription, and to metastatic development.24,25 A third line of evidence suggests that PSF could help to transform normal differentiated cells into tumor cells via mutations in PSF that could reduce PSF abundance or PSF localization, as seen for the fusion protein PSF-TFE3 (transcription factor binding to IGHM enhancer 3), which is found in a subset of renal cell carcinomas.26,27 Fourth, PSF interacts with RNF43, another oncogenic RING finger protein that has the ability to stimulate cell growth. As it was proposed that RNF43 exerted a growth-promoting influence, the heterodimer RNF43-PSF could also impact upon diverse nuclear events relevant to tumorigenesis.28
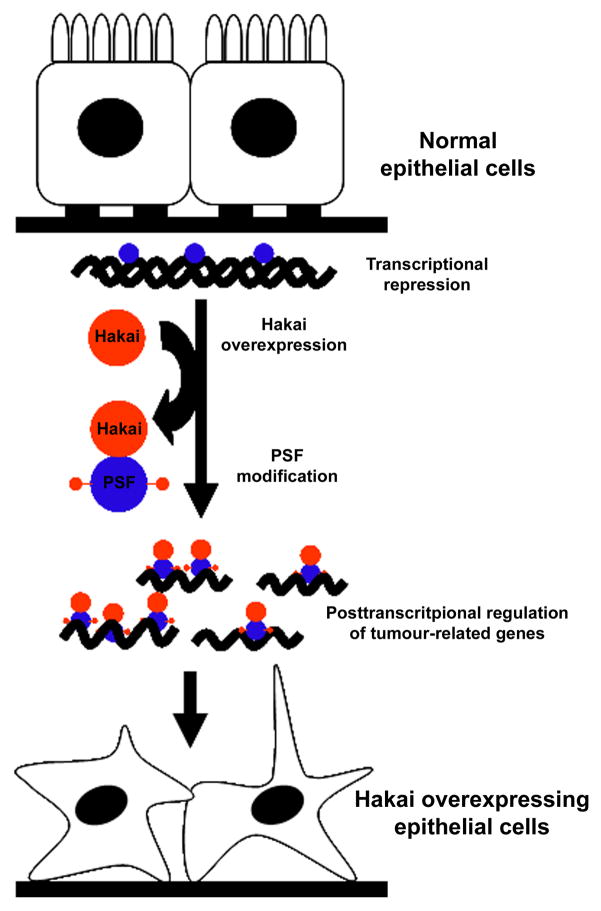
Taken together, we hypothesize that PSF can act as a repressor of oncogenic genes through its DNA-binding properties; under specific physiological or pathological conditions, such as when Hakai is overexpressed (Figure 2), PSF could revert its tumor suppressive action by detaching from DNA, associating instead with mRNA molecules, post-transcriptionally regulating the fate of mRNAs targets, and thereby promoting tumorigenesis. As we elucidate further details of this working model, we will gain a deeper understanding of the role of Hakai and PSF in epithelial tumorigenesis.
Figure 2. Diagram of PSF function as DNA-binding protein in normal cells and as RNA-binding protein in Hakai-overexpressing cells.
In normal epithelial cells, PSF is associated with DNA and represses cancer-related gene expression. Under conditions of Hakai overexpression, PSF is post-translationally modified, which turns it into an RNA-binding protein with higher affinity for many mRNAs encoding cancer-promoting proteins.
Acknowledgments
A.F. is supported by Parga Pondal Program, Secretaria Xeral I+D+I, Xunta de Galicia, Spain. Y.F. is supported by MRC funding to the Cell Biology Unit. M.G. is supported by the NIA-IRP, NIH.
Abbreviations
- BRK
breast tumor kinase
- EGF
epidermal growth factor
- HGF
hepatocyte growth factor
- Mnk
mitogen-activated protein kinase signal-integrating kinase
- NPM/ALK
tyrosine kinase nucleophosmin/anaplastic lymphoma kinase
- PSF
polypyrimidine tract-binding-protein-associated splicing factor
- RRM
RNA recognition motif
References
- 1.Fujita Y, Krause G, Scheffner M, Zechner D, Leddy HE, Behrens J, Sommer T, Birchmeier W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol. 2002;4:222–31. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- 2.Palacios F, Tushir JS, Fujita Y, D’Souza-Schorey C. Lysosomal targeting of E-cadherin: a unique mechanism for the down-regulation of cell-cell adhesion during epithelial to mesenchymal transitions. Mol Cell Biol. 2005;25:389–402. doi: 10.1128/MCB.25.1.389-402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen Y, Hirsch DS, Sasiela CA, Wu WJ. CDC42 regulates E-cadherin ubiquitination and degradation through an EGF receptor to SRC-mediated pathway. J Biol Chem. 2007;283:5127–37. doi: 10.1074/jbc.M703300200. [DOI] [PubMed] [Google Scholar]
- 4.Bonazzi M, Veiga E, Cerda JP, Cossart P. Successive post-translational modifications of E-cadherin are required for InlA-mediated internalisation of Listeria monocytogenes. Cell Microbiol. 2008;10:2208–22. doi: 10.1111/j.1462-5822.2008.01200.x. [DOI] [PubMed] [Google Scholar]
- 5.Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806–11. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 6.Birchmeier W, Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta. 1994;1198(1):11–26. doi: 10.1016/0304-419x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 7.Perez-Moreno M, Jamora C, Fuchs E. Sticky business: orchestrating cellular signals at adherens junctions. Cell. 2003;112:535–548. doi: 10.1016/s0092-8674(03)00108-9. [DOI] [PubMed] [Google Scholar]
- 8.Wheelock MJ, Johnson KR. Cadherin-mediated cellular signaling. Curr Opin Cell Biol. 2003;15:509–514. doi: 10.1016/s0955-0674(03)00101-7. [DOI] [PubMed] [Google Scholar]
- 9.Pece S, Gutkind JS. E-cadherin and Hakai: signalling, remodeling or destruction? Nat Cell Biol. 2002;4:E72–4. doi: 10.1038/ncb0402-e72. [DOI] [PubMed] [Google Scholar]
- 10.Figueroa A, Kotani H, Toda H, Mazan-Mamczarz K, Mueller EC, Otto A, Disch L, Keshtgar M, Gorospe M, Fujita Y. Novel roles of Hakai in cell proliferation and oncogenesis. Mol Biol Cell. 2009;20:3533–42. doi: 10.1091/mbc.E08-08-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 12.Basu A, Dong B, Krainer AR, Howe CC. The intracisternal A-particle proximal enhancer-binding protein activates transcription and is identical to the RNA- and DNA-binding protein p54nrb/NonO. Mol Cell Biol. 1997;17:677–86. doi: 10.1128/mcb.17.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathur M, Tucker PW, Samuels HH. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol. 2001;21:2298–311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zolotukhin AS, Michalowski D, Bear J, Smulevitch SV, Traish AM, Peng R, Patton J, Shatsky IN, Felber BK. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol Cell Biol. 2003;23:6618–30. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong X, Sweet J, Challis JR, Brown T, Lye SJ. Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol Cell Biol. 2007;27:4863–75. doi: 10.1128/MCB.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–89. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol Cell Biol. 2007;27:6972–84. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, Lilley KS, Bushell M, Willis AE. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol Cell Biol. 2008;28:40–9. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 20.Dye BT, Patton JG. An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp Cell Res. 2001;263:131–44. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]
- 21.Buxadé M, Morrice N, Krebs DL, Proud CG. The PSF. p54nrb complex is a novel Mnk substrate that binds the mRNA for tumor necrosis factor alpha. J Biol Chem. 2008;283:57–65. doi: 10.1074/jbc.M705286200. [DOI] [PubMed] [Google Scholar]
- 22.Galietta A, Gunby RH, Redaelli S, Stano P, Carniti C, Bachi A, Tucker PW, Tartari CJ, Huang CJ, Colombo E, Pulford K, Puttini M, Piazza RG, Ruchatz H, Villa A, Donella-Deana A, Marin O, Perrotti D, Gambacorti-Passerini C. NPM/ALK binds and phosphorylates the RNA/DNA-binding protein PSF in anaplastic large-cell lymphoma. Blood. 2007;110:2600–9. doi: 10.1182/blood-2006-01-028647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukong KE, Huot ME, Richard S. BRK phosphorylates PSF promoting its cytoplasmic localization and cell cycle arrest. Cell Signal. 2009;21:1415–22. doi: 10.1016/j.cellsig.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Song X, Sui A, Garen A. Binding of mouse VL30 retrotransposon RNA to PSF protein induces genes repressed by PSF: effects on steroidogenesis and oncogenesis. Proc Natl Acad Sci USA. 2004;101:621–6. doi: 10.1073/pnas.0307794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song X, Sun Y, Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc Natl Acad Sci U S A. 2005;102:12189–93. doi: 10.1073/pnas.0505179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, Hamoudi R, Linehan WM, Shipley J, Cooper CS. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–9. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 27.Mathur M, Das S, Samuels HH. PSF-TFE3 oncoprotein in papillary renal cell carcinoma inactivates TFE3 and p53 through cytoplasmic sequestration. Oncogene. 2003;22:5031–44. doi: 10.1038/sj.onc.1206643. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto K, Sakurai H, Sugiura T. Proteomic identification of a PSF/p54nrb heterodimer as RNF43 oncoprotein-interacting proteins. Proteomics. 2008;8:2907–10. doi: 10.1002/pmic.200800083. [DOI] [PubMed] [Google Scholar]