Abstract
Abnormalities of fatty acid metabolism are recognized to play a significant role in human disease, but the mechanisms remain poorly understood. Long-chain acyl-CoA dehydrogenase (LCAD) catalyzes the initial step in mitochondrial fatty acid oxidation (FAO). We produced a mouse model of LCAD deficiency with severely impaired FAO. Matings between LCAD +/− mice yielded an abnormally low number of LCAD +/− and −/− offspring, indicating frequent gestational loss. LCAD −/− mice that reached birth appeared normal, but had severely reduced fasting tolerance with hepatic and cardiac lipidosis, hypoglycemia, elevated serum free fatty acids, and nonketotic dicarboxylic aciduria. Approximately 10% of adult LCAD −/− males developed cardiomyopathy, and sudden death was observed in 4 of 75 LCAD −/− mice. These results demonstrate the crucial roles of mitochondrial FAO and LCAD in vivo.
Mitochondrial fatty acid oxidation (FAO) is the primary means by which energy is derived from metabolism of fatty acids. This process is important during periods of fasting or prolonged strenuous activity, providing as much as 80 to 90% of fatty acid-derived energy for heart and liver function (1). Mitochondrial FAO also provides acetyl-CoA for hepatic ketogenesis and the energy required for nonshivering thermogenesis by brown adipose tissue (2). The initial step in mitochondrial FAO is the α-β dehydrogenation of the acyl-CoA ester by a family of four closely related, chain length-specific enzymes, the acyl-CoA dehydrogenases, which include very-long-chain, long-chain, medium-chain, and short-chain acyl-CoA dehydrogenases (VLCAD, LCAD, MCAD, and SCAD, respectively). These enzymes catalyze the same type of reaction but differ in specificity according to the chain length of their fatty acid (acyl-CoA) substrates.
Human diseases caused by deficiencies in each of these enzymes except LCAD have been reported, with MCAD deficiency having the highest prevalence (3). Patients affected with any one of these deficiencies present with a wide range of clinical phenotypes ranging from lethargy and muscle weakness to severe liver disease and potentially fatal cardiomyopathy. MCAD deficiency has been implicated in cases of sudden infant death (4). Specific mutations that cause disease have been identified in the genes encoding human SCAD (ACADS; ref. 5), MCAD (ACADM; ref. 6), and VLCAD (ACADVL; ref. 7). Although human patients have been reported with putative LCAD deficiency (8), they were diagnosed before the discovery of VLCAD (9), and most of them were found later to be VLCAD deficient (10). Thus far, no specific mutations have been identified in the gene encoding human LCAD (ACADL), and no definitively diagnosed cases of human LCAD deficiency are known. The failure to identify patients with LCAD deficiency is surprising given the recognition of disease caused by deficiencies in all other members of this gene family. This has put into question both the role of LCAD in the metabolism of long-chain fatty acids and the potential of LCAD deficiency to produce disease. Human LCAD deficiency may not cause clinical disease or it could result in gestational lethality, either of which could account for the lack of identified human cases. To evaluate the functional role of LCAD in mitochondrial FAO in vivo, we developed a mouse model of LCAD deficiency by targeted mutagenesis of the gene encoding LCAD (Acadl) in mouse embryonic stem (ES) cells.
MATERIALS AND METHODS
Generation of LCAD-Deficient Mice.
The targeting vector pAcadltm1Uab was constructed by using a 7.5-kb Acadl (NotI/HindIII) fragment of 129/SvJ DNA and a neor cassette derived from PGKneobpA (11), under the control of the phosphoglycerate kinase gene promoter and a bovine poly(A) signal and subcloned into pGEM-11zf(+) (Promega). An 821-bp deletion of the Acadl sequence, spanning exon 3 with flanking intron sequence, was created in the vector before electroporation and served as the site of linearization. Repair of this deletion on homologous recombination via the double-stranded-break repair model (12) served as the basis for screening ES cell colonies for correct targeting by Southern blot analysis. Duplication of exon 3 can occur only on homologous recombination. Linearized vector was electroporated into TC-1 ES cells (13) derived from 129/SvEvTacfBR (129) mice, and G418-resistant clones were analyzed by using Southern blot analysis. Correctly targeted clones were microinjected into C57BL/6J (B6) blastocysts to generate chimeras that were backcrossed to C57BL/6NTacfBR mice (Taconic). All mice analyzed in these studies were generation 2–3 with B6,129-Acadltm1Uab/tm1Uab (LCAD −/−) or B6,129-Acadl+/+ (normal control) genotypes from intercrosses of B6,129-Acadltm1Uab/+ (LCAD −/+) mice. Genotypes were determined by using Southern blot analysis. Mice were negative for murine pathogens based on a panel of 10 virus serologies, aerobic bacterial cultures of nasopharynx and cecum, endo- and ectoparasite exams, and histopathology of all major organs. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham.
RNA Analysis.
Total RNA from liver, heart, skeletal muscle, kidney, and brain was isolated from 21-to-28-day-old mice by using the guanidinium thiocyanate method (14). First-round cDNA synthesis was performed by using 1–2 μg of total RNA from heart and random primers as recommended by the manufacturer (Invitrogen). PCR was performed by using Acadl-specific primers to amplify the first 5 exons of the Acadl+ or Acadltm1Uab transcripts (5′-ATGGCTGCG CGCCTGCTCCTC and 3′-CTTGCTTCCATTGAGAATCCA). After an initial 4.5-min, 94°C denaturation, 32 cycles were performed at 94°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min. Products were visualized by electrophoresis on a 2% agarose gel. PCR products from Acadl+ or Acadltm1Uab transcripts were subcloned into pGEM-T Easy vector (Promega) for sequence analysis.
Enzymology.
Liver mitochondria were isolated from 4- to 5-wk-old mice, and immunoblot analysis was performed on 50 μg of liver mitochondrial protein as described (15) by using rat, anti-LCAD antibodies. Enzyme activity toward palmitoyl-CoA (C16:0) and octanoyl-CoA(C8:0) was determined on extracts from cultured fibroblasts by using the electron transport flavoprotein fluorescence-reduction assay (16).
Biochemical Analysis.
Serum glucose analysis was performed by using the glucose oxidase method (Sigma). Urine organic acids, as well as serum and tissue free fatty acids (FFA), were analyzed by using GC-MS (17, 18). Bile acylcarnitine analysis was performed by using electrospray tandem mass spectrometry (19). Whole blood spots and skeletal muscle were analyzed for specific acylcarnitine derivatives by using fast-atom bombardment–tandem mass spectrometry (20).
Statistical Analysis.
Statistical analysis was performed by using a one-way ANOVA for comparison of the means of measured values. Paired-means comparison was done by using the Tukey method with P < 0.05 accepted as significant. Genotype distribution of pups from LCAD heterozygous crosses was analyzed by the χ2 test for variance from expected results. All statistical analyses were performed using the statistix 4.0 program (Analytical Software).
RESULTS
Targeting of Acadl in Mice.
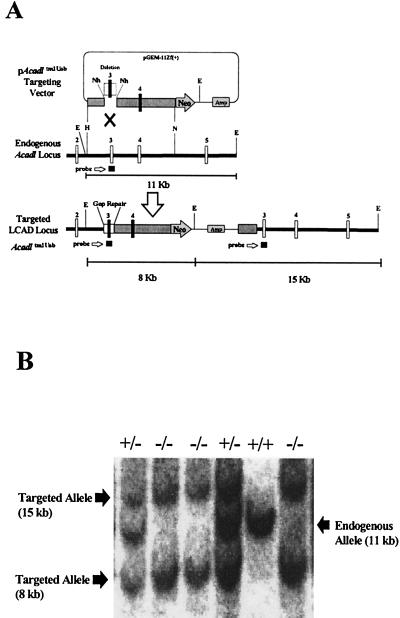
A 7.5-kb HindIII/NotI fragment of Acadl (21) containing exons 3 and 4 with flanking intron sequence along with a neor gene was used as an insertion vector for homologous recombination in TC-1 mouse ES cells (Fig. 1A). The insertion created a duplication of exons 3 and 4 separated by the neor gene and plasmid sequence. Two cell lines with the correct targeting were identified by using Southern blot analysis. One of these lines produced 10 chimeric mice, of which 5 were found to pass the mutant allele (Acadltm1Uab) to offspring. The heterozygous offspring (B6, 129-Acadl+/tm1Uab) were then intercrossed to produce the three genotypes: LCAD +/+ (B6, 129-Acadl+/+), LCAD +/− (B6, 129-Acadl+/tm1Uab), and LCAD −/− (B6, 129-Acadltm1Uab/tm1Uab) (Fig. 1B).
Figure 1.
Targeting of the mouse Acadl gene. (A) Schematic diagram of the targeting vector, pAcadltm1Uab, the endogenous Acadl locus, and the targeted Acadltm1Uab locus. Gap repair of the deleted region (an 821-bp NheI deletion of exon 3 and flanking intron sequence) in the targeting vector occurs on homologous recombination and was the basis for genotype screening by Southern blot analysis of EcoRI-digested DNA. E, EcoRI; H, HindIII; N, NotI; Nh, NheI. (B) Representative Southern blot from Acadl-targeted mice showing three genotypes. EcoRI-digested genomic DNA was probed with a 304-bp Acadl genomic fragment containing exon 3 and flanking intron sequence. Banding patterns are as follows: Acadl normal controls (+/+), −11 kb; heterozygous (+/−), −15 kb, 11 kb, and 8 kb; and homozygous mutant (−/−), −15 kb and 8 kb.
RNA Analysis.
Northern blot analysis of total RNA isolated from liver, heart, skeletal muscle, kidney, and brain demonstrated the two prominent bands as described (22). There were no detectable differences observed from LCAD −/−, +/−, and +/+ mice (data not shown). Reverse transcriptase-PCR analysis of total RNA from heart, amplifying the first five exons of the Acadl+ or Acadltm1Uab transcripts from LCAD −/−, +/−, or +/+ mice, demonstrated a single product from the +/+ and −/− mice (604 bp vs. 907 bp, respectively) and both products (604 and 907 bp) from the +/− mice (data not shown). Analysis of the wild-type (+/+) cDNA demonstrated the expected normal mRNA sequence as reported (22), and the cDNA from the targeted allele (Acadltm1Uab) demonstrated an exact duplication of exons 3 and 4 with removal of the neor and plasmid sequence on splicing (data not shown). The duplication of exons 3 and 4 in the mRNA did not disrupt the reading frame with premature stop codons. However, in proteins isolated from liver mitochondria of LCAD −/− mice, no LCAD could be detected by immunoblotting with rat anti-LCAD antibody (Fig. 2).
Figure 2.
LCAD immunoblot. Immunoblot of liver mitochondrial proteins (50 μg) isolated from normal control (+/+), heterozygous mutant (+/−), and homozygous mutant (−/−) mice and probed with a rat anti-LCAD antibody. No detectable LCAD protein was observed in samples from the homozygous mutant mice.
Gestational Loss.
Matings of LCAD +/− mice did not produce the expected number of progeny. There were 500 pups born (87 litters = 5.8 ± 3.5 pups per litter) compared with LCAD +/+ controls (12 litters = 8.1 ± 2.8 pups per litter). There were 479/500 (95.8% of total born) pups genotyped (LCAD +/+ = 192; LCAD +/− = 173; LCAD −/− = 114), and 434 of them (85.6% of total born) were weaned. Analyzing the results based on the number genotyped with an expected 1:2:1 (LCAD +/+:+/−:−/−) distribution, we find that these results deviate significantly from the expected (P < 0.001), indicating an abnormally low number of LCAD +/− and an abnormally high number of LCAD +/+ phenotypes. There were, however, significantly lower numbers of pups per litter (P < 0.02) in the heterozygous matings than in wild-type controls (5.8 vs. 8.1). Alternatively, if we assume that the number of LCAD +/+ offspring represents the expected number for a 25% distribution, then we would expect a total of 768 pups born from the 87 litters, or 8.8 pups per litter, which is closer to the number actually observed (8.1 pups per litter) in control matings. By using this assumption, the distribution deviates significantly from the expected (P < 0.001), with a lower number of LCAD +/− and LCAD −/− pups born, which would account for the small litter size found, i.e., [LCAD +/+ (192 expected:192 observed (100%)], LCAD +/− [384 expected: 173 observed (45%)] and LCAD −/− [192 expected: 114 observed (60%)]. These breeding pairs were closely observed by three different individuals at least twice per day, and any neonatal deaths were accounted for in the results described here.
Enzymology.
The degree of functional overlap in chain-length specificity of the four acyl-CoA dehydrogenase enzymes has caused considerable difficulty in the diagnosis of human acyl-CoA dehydrogenase deficiencies based on biochemical and enzymatic data, and affected patients often cannot be given a specific diagnosis. Extracts from cultured fibroblasts of LCAD −/− mice demonstrated a 28% reduction in ability to dehydrogenate palmitoyl-CoA (C16:0) in comparison to extracts from LCAD +/+ fibroblasts (Table 1). The residual activity in the LCAD −/− extracts toward palmitoyl-CoA was the result of endogenous VLCAD activity and could be abolished by preincubation with anti-VLCAD antibodies, indicating the LCAD-specific activity was undetectable.
Table 1.
Dehydrogenation of palmitoyl-CoA (C16:0) and octanoyl-CoA (C8:0) using the electron transport flavoprotein reduction fluorescence assay in cellular extracts from cultured fibroblasts
| LCAD Genotype | Substrate | Antibody | Specific activity, milliunit/mg protein | Wild-type activity, % |
|---|---|---|---|---|
| +/+ | Palmitoyl-CoA | None | 2.17 ± 0.005 | 100 |
| Palmitoyl-CoA | Anti-VLCAD | 1.34 ± 0.09 | 62* | |
| Octanoyl-CoA | None | 2.90 | ||
| +/− | Palmitoyl-CoA | None | 2.45 ± 0.10 | 110 |
| Palmitoyl-CoA | Anti-VLCAD | 1.60 ± 0.04 | 65 | |
| Octanoyl-CoA | None | 2.32 | ||
| −/− | Palmitoyl-CoA | None | 1.56 ± 0.11 | 72 |
| Palmitoyl-CoA | Anti-VLCAD | Not detectable | 0 | |
| Octanoyl-CoA | None | 2.16 |
Values reported are mean ± SD, n = 3. *, % Activity with no VLCAD antibody.
Fasting Intolerance.
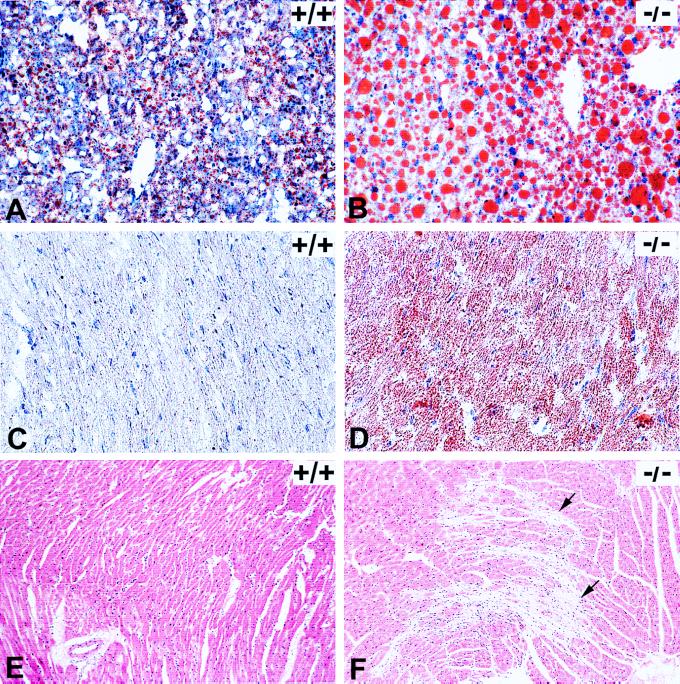
After an overnight fast (18–20 hr), LCAD −/− mice developed severe and diffuse micro- and macrovesicular hepatic steatosis (Figs. 3 A and B) as well as microvesicular cardiac lipidosis (Figs. 3 C and D). The fasted LCAD −/− mice had significantly (P < 0.005) reduced mean serum glucose concentration (78.9 ± 59.7 mg/dl) in contrast to fasted normal controls (147.7 ± 95.3 mg/dl). The serum glucose concentrations in the LCAD −/− mice were highly variable as is often seen in patients with other FAO disorders (3). Serum FFA analysis of fasted mice demonstrated a significant elevation (P < 0.05) in total FFAs in the LCAD −/− mice as compared with fasted normal controls, with specific elevations in unsaturated fatty acids: docenedioic (C12:1), tetradecenedioic (C14:1), tetradecadienoic (C14:2), oleic (C18:1), and linoleic (C18:2) acids (Table 2). Liver FFA analysis showed no significant differences in total FFA, but elevations of C10–C14 fatty acids were observed in the fasted LCAD −/− mice (Table 2).
Figure 3.
Histopathology on LCAD +/+ (normal control) and LCAD −/− (deficient mice). (A) +/+ and (B) −/−: Oil-Red-O staining of frozen liver sections from 4- to 5-wk-old mice after an 18- to 20-hr fast. Abundant large lipid droplets present in the LCAD-deficient mice compared with minimal lipid accumulation in normal control. (C) +/+ and (D) −/−: Oil-Red-O staining of frozen hearts from 14- to 16-wk-old mice after an 18- to 20-hr fast. Note lipid accumulation in all cardiomyocytes of the LCAD-deficient mouse as compared with the LCAD-normal control. (E) +/+ and (F) −/−: Hematoxylin/eosin-stained heart sections from 16-wk-old, nonfasted male mice. Note prominent myocardial degeneration and fibrosis (arrows) present in the LCAD −/− mice (F). (A and B, ×25, C and D, ×40, and E and F, ×10.)
Table 2.
Mean serum and tissue free fatty acids and acylcarnitine metabolites
| Fatty acid | Serum-free fatty acids, μmol/liter | Liver fatty acids, mmol/mg protein | Whole blood acyl-carnitine, nmol/ml | Skeletal muscle acyl-carnitine, nmol/ml | ||||
|---|---|---|---|---|---|---|---|---|
| +/+ | −/− | +/+ | −/− | +/+ | −/− | +/+ | −/− | |
| C8 | 9.50 | 7.60 | 0.010 | 0.046 | 0.010 | 0.050 | 0.066 | 0.043 |
| C10:1 | 0.400 | 0.400 | 0.001 | 0.001 | 0.020 | 0.040 | 0.017 | 0.009 |
| C10 | 5.30 | 4.20 | 0.005 | 0.047* | 0.010 | 0.050 | 0.037 | 0.012 |
| C12:1 | 0.100 | 1.00* | 0.000 | 0.060* | 0.450 | 1.40* | 0.026 | 0.053 |
| C12 | 6.90 | 8.20 | 0.016 | 0.387* | 0.550 | 1.60 | 0.021 | 0.078 |
| C14:2 | 2.40 | 11.0* | 0.033 | 0.225* | 0.160 | 5.91* | 0.022 | 0.124* |
| C14:1 | 1.20 | 26.8* | 0.005 | 0.110* | 0.580 | 13.9* | 0.067 | 0.237* |
| C14 | 25.9 | 22.4 | 0.084 | 0.170* | 1.07 | 1.80 | 0.107 | 0.082 |
| C16:1 | 15.0 | 32.0 | 0.200 | 0.533* | — | — | — | — |
| C16 | 240 | 325 | 3.97 | 4.57 | 2.16 | 4.82 | 0.583 | 0.388 |
| C18:2 | 429 | 882* | 4.37 | 5.73 | 0.780 | 2.06 | 0.178 | 0.114 |
| C18:1 | 199 | 374* | 3.00 | 3.87 | 0.920 | 1.87 | 0.397 | 0.260 |
| C18 | 305 | 312 | 1.03 | 1.30 | — | — | — | — |
| Total | 1239 | 2007* | 12.6 | 16.5 | — | — | — | — |
All mice (n = 3 for both groups) were 4–5 wk old and fasted for 18–20 hr. *P < 0.05; —, not measured.
Organic Acid and Carnitine Analyses.
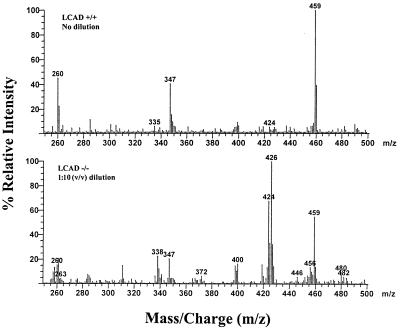
The initial diagnosis of long-chain FAO disorders is made most often by analyzing a variety of biochemical markers such as organic acids and acylcarnitines. Urine organic acid analysis of fasted LCAD −/− mice demonstrated excretion of high levels of adipic (C6:0), octenedioic (C8:1), decenedioic (C10:1), and hydroxydecenedioic (C10:1) acids compared with fasted normal controls (data not shown). In particular, high levels of octenedioic acid have not been observed in the urine of patients with other FAO disorders and may represent a possible distinctive feature of LCAD deficiency. Acylcarnitine analysis of whole blood and skeletal muscle from fasted LCAD −/− mice demonstrated elevations of C12–C14 acylcarnitines with a predominance of tetradecenoyl-(C14:1) and tetradecadienoyl-(C14:2) carnitines compared with fasted normal controls (Table 2). Long-chain acylcarnitines also have been observed in postmortem bile samples from patients with FAO defects (19). In bile from fasted LCAD −/− mice, we detected extremely high concentrations of C14:1 and C14:2 acylcarnitines (Fig. 4) with values exceeding 1 mM (fasted, normal controls: 0.05–0.07 mM). These results indicate that a major route of excretion for long-chain fatty acids is the bile and confirm the importance of bile acylcarnitine profiles in the diagnosis of FAO disorders (19).
Figure 4.
Butyl-ester acylcarnitine profiles of bile specimens obtained by using precursor-ion scanning (parent of m/z 85) electrospray/tandem mass spectrometry. Mice were sacrificed after overnight fast (18–20 hr), and bile specimens were collected as spots on filter paper. (Top) LCAD +/+ (normal control), no dilution. (Bottom) LCAD −/− (LCAD-deficient), 1:10 (vol/vol) dilution. Internal standards: m/z 263, d3-acetylcarnitine; m/z 347, d3-octanoylcarnitine; and m/z 459, d3-palmitoylcarnitine. Peak identification: m/z 260, acetylcarnitine (C2); m/z 372, decanoylcarnitine (C10); m/z 400, dodecanoylcarnitine (C12); m/z 424, tetradecadienoylcarnitine (C14:2); m/z 426, tetradecenoylcarnitine (C14:1); m/z 456, palmitoylcarnitine (C16); m/z 480, linoleylcarnitine (C18:2); m/z 482, oleylcarnitine (C18:1).
Sudden Death and Cardiac Lesions.
Surprisingly, we observed sudden death in 4 phenotypically normal, nonfasted, LCAD −/− mice of a total of 75 mutants used in these studies. Two of these mice were 4–5 weeks of age, and 2 were 14 weeks of age. These mice died suddenly in the absence of undue stress from handling or other environmental conditions. In histopathologic examination of mice presented in these studies, microvesicular fatty change in the heart of 4- to 5-wk-old fasted LCAD −/− mice was the only cardiac lesion noted in this age group. In adult (14–16-wk-old) LCAD −/− mice, fasting produced similar cardiac and hepatic fatty change as seen in 4- to 5-wk-old LCAD −/− mice. Additionally, 10% of adult (14 to 16 week old) LCAD −/− males were found to have severe, multifocal myocardial fibrosis, suggesting cardiomyocyte degeneration with replacement by connective tissue (Fig. 3F). These lesions were not seen in age-matched normal control mice (Fig. 3E) or LCAD −/− female mice.
DISCUSSION
Currently, there is much interest in the role of fatty acids in human diseases including obesity, diabetes, insulin resistance, cancer, and cardiovascular disease (23, 24, 25). Thus far, no animal model has existed with a specific genetic defect of long-chain FAO to evaluate this pivotal component in vivo. Long-chain fatty acids are the primary fatty acids found in human and other animal diets and the ultimate components of stored body fat.
One of the first striking results found in these mice was the gestational loss of both LCAD +/− and −/− pups. The metabolic cavitation model (26) proposed that mitochondria in the outer blastomeres colocalize with lipid droplets to the basolateral aspects for β-oxidation of fatty acids, e.g., palmitic acid. This would supply the required ATP for Na+/K+ ATPase activity essential for producing the nascent blastocoel fluid. Therefore, one possible explanation in LCAD deficiency would be that FAO is required during blastocyst formation, and this process is insufficient in some LCAD +/− or −/− embryos, causing them to die very early in gestation. Alternatively, severe maternal complications like acute fatty liver of pregnancy and HELLP (hemolysis, elevated liver enzymes, and low platelets) syndrome have been reported in women who were heterozygous deficient for another enzyme involved in mitochondrial FAO, long-chain 3-hydroxyacyl-CoA dehydrogenase (LCHAD) and carrying an LCHAD−/− fetus (27). The mechanism of the hepatocellular damage in this disorder is still unclear, but is likely to include the accumulation by the fetus of abnormal fatty acid metabolites that enter the maternal circulation and overwhelm the mitochondrial oxidation pathway of the heterozygous mother. Similarly, in the LCAD +/− mouse carrying several LCAD −/− offspring, it appears feasible that a metabolic environment may exist that is detrimental to the development of LCAD +/− and −/− fetuses. Finally, these effects conceivably could result from the influences of unknown background genetic factors. Clearly, the mechanism involved in this finding is unknown and requires further investigation.
The enzymology of the LCAD-deficient mice demonstrates clearly an antigen-negative phenotype with severely impaired oxidation of fatty acids as shown in fibroblasts. In contrast, cultured human fibroblasts demonstrated dehydrogenation activity toward palmitoyl-CoA that was >97% attributable to VLCAD activity (7). Additionally, recent evidence has shown that human LCAD has significant activity toward branched long-chain substrates (28). Our data may point to a species variation in the substrate specificity of LCAD and VLCAD, as has been demonstrated with purified pig LCAD and recombinant human LCAD (29) or to differences between mouse and man in the absolute expression of LCAD and VLCAD.
LCAD −/− mice that achieve birth appear normal, as do most children with other acyl-CoA dehydrogenase deficiencies. In such human patients, clinical disease is most often exacerbated during fasting, when FAO is needed to maintain energy homeostasis. Affected patients often present with hypoglycemia, hypoketonemia, elevated serum free fatty acids, and nonketotic organic aciduria. LCAD −/− mice developed macroscopic evidence of a fatty liver that is visible grossly after only 4 hr of fasting, compared with no gross changes in normal controls. We witnessed sudden death in the LCAD −/− mice during conditions of no apparent external stress. In contrast, we have never seen sudden death in the BALB/cByJ mouse model of SCAD deficiency (17) nor in the LCAD +/+ or +/− mice. Patients with VLCAD deficiency can develop cardiac arrhythmias (30) and cardiomyopathy (31), resulting in death at an early age. Although the exact cause of these cardiac changes was unknown, the elevated concentrations of long-chain acylcarnitines observed in these patients are known to be arrhythmogenic (25). Animal studies also have shown that pharmacological inhibition of long-chain FAO by 2-tetradecylglycidate leads to the development of cardiomyopathy (32).
These studies indicate that LCAD has an important role in mitochondrial FAO and the maintenance of fasting energy homeostasis. LCAD deficiency results in gestational loss, severe fasting intolerance with subsequent hepatic and cardiac lipidosis, hypoglycemia, elevated serum FFAs, nonketotic dicarboxylic aciduria, and myocardial degeneration. Although the data suggest that LCAD may have a greater role in mitochondrial FAO in mice than in humans, the complete LCAD deficiency found in this mouse model can be used to predict the phenotype of putative human LCAD deficiency. Moreover, the LCAD-deficient mouse develops a clinical syndrome virtually identical to human VLCAD deficiency. These studies provide strong evidence that mitochondrial FAO in the mouse has crucial roles in embryonic/fetal development, fasting energy homeostasis, and cardiac function and that LCAD is essential for these functions.
Acknowledgments
We thank Susan C. Farmer for her early contributions to our ES cell work and review of the manuscript, Chuxia Deng and Philip Leder for the TC-1 ES cells, and Phillippe Soriano for the PGKneobpA plasmid. We gratefully acknowledge support by the University of Alabama at Birmingham-Comprehensive Cancer Center Core facilities (National Cancer Institute Grant P30CA-13148) including the Transgenic Animal/Embryonic Stem Cell Resource (C.A.P., Director) and the Oligonucleotide Facility (J. Engler, Director), and the DNA Sequencing Facility supported in part by the Howard Hughes Medical Institute (S. Hollingshead, Director). This work was supported by National Institutes of Health Grants RO1-RR02599 (P.A.W.) and RO1-DK45482 (J.V.).
ABBREVIATIONS
- ES
embryonic stem
- FAO
fatty acid oxidation
- LCAD
long-chain acyl-CoA dehydrogenase
- Acadl
symbol for mouse gene encoding LCAD
- VLCAD
very long-chain AD
- MCAD
medium-chain AD
- SCAD
short-chain AD
- FFA
free fatty acids
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Mannaerts G P, Debeer L J. Ann NY Acad Sci. 1982;386:30–39. doi: 10.1111/j.1749-6632.1982.tb21405.x. [DOI] [PubMed] [Google Scholar]
- 2.Nicholls D G, Locke R M. Physiol Rev. 1984;64:1–64. doi: 10.1152/physrev.1984.64.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Roe C R, Coates P M. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 1501–1533. [Google Scholar]
- 4.Boles R G, Buck E A, Blitzer M G, Platt M S, Cowan T M, Martin S K, Yoon H, Madsen J A, Reyes-Mugica M, Rinaldo P. J Pediatr (Berlin) 1998;132:924–933. doi: 10.1016/s0022-3476(98)70385-3. [DOI] [PubMed] [Google Scholar]
- 5.Naito E, Ozasa H, Ikeda Y, Tanaka K. J Clin Invest. 1989;83:1605–1613. doi: 10.1172/JCI114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokota I, Indo Y, Coates P M, Tanaka K. J Clin Invest. 1990;86:1000–1003. doi: 10.1172/JCI114761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aoyama T, Souri M, Ueno I, Kamijo T, Yamaguchi S, Rhead W J, Tanaka K, Hashimoto T. Am J Hum Genet. 1995;57:273–283. [PMC free article] [PubMed] [Google Scholar]
- 8.Hale D E, Stanley C A, Coates P M. In: Fatty Acid Oxidation: Clinical, Biochemical, and Molecular Aspects. Tanaka K, Coates P M, editors. New York: Liss; 1990. pp. 303–324. [Google Scholar]
- 9.Izai K, Uchida Y, Orii T, Yamamoto S, Hashimoto T. J Biol Chem. 1992;267:1027–1033. [PubMed] [Google Scholar]
- 10.Yamaguchi S, Indo Y, Coates P M, Hashimoto T, Tanaka K. Pediatr Res. 1993;34:111–113. doi: 10.1203/00006450-199307000-00025. [DOI] [PubMed] [Google Scholar]
- 11.Soriano P, Montgomery C, Geske R, Bradley A. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 12.Scostak J W, Orr-Weaver T L, Rothenstein R J. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 13.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 14.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Amendt B A, Freneaux E, Reece C, Wood P A, Rhead W J. Pediatr Res. 1992;31:552–556. doi: 10.1203/00006450-199206000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Mohsen A W, Vockley J. Biochemistry. 1995;34:10146–10152. doi: 10.1021/bi00032a007. [DOI] [PubMed] [Google Scholar]
- 17.Wood P A, Amendt B A, Rhead W J, Millington D S, Inoue F, Armstrong D. Pediatr Res. 1989;25:38–43. doi: 10.1203/00006450-198901000-00010. [DOI] [PubMed] [Google Scholar]
- 18.Boles R G, Martin S K, Blitzer M G, Rinaldo P. Hum Pathol. 1994;25:735–741. doi: 10.1016/0046-8177(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 19.Rashed M S, Ozand P T, Bennett M J, Barnard J J, Govindaraju D R, Rinaldo P. Clin Chem. 1995;41:1109–1114. [PubMed] [Google Scholar]
- 20.Millington D S, Norwood D L, Kodo N, Roe C R, Inoue F. Anal Biochem. 1989;180:331–339. doi: 10.1016/0003-2697(89)90441-7. [DOI] [PubMed] [Google Scholar]
- 21.Kurtz D M, Tolwani R J, Wood P A. Mamm Genome. 1998;9:361–365. doi: 10.1007/s003359900770. [DOI] [PubMed] [Google Scholar]
- 22.Hinsdale M E, Farmer S C, Johnson K R, Davisson M T, Hamm D A, Tolwani R J, Wood P A. Genomics. 1995;28:163–170. doi: 10.1006/geno.1995.1127. [DOI] [PubMed] [Google Scholar]
- 23.Guenther B. Diabetes. 1997;46:3–10. [Google Scholar]
- 24.Welsch C W. FASEB J. 1991;5:2160–2166. [Google Scholar]
- 25.Oliver M F, Opie L H. Lancet. 1994;343:155–158. doi: 10.1016/s0140-6736(94)90939-3. [DOI] [PubMed] [Google Scholar]
- 26.Wiley L M. In: The Mammalian Preimplantation Embryo - Regulation of Growth and Differentiation in Vitro. Bavister B D, editor. New York: Plenum; 1987. pp. 65–93. [Google Scholar]
- 27.Treem W R, Shoup M E, Hale D E, Bennet M J, Rinaldo P, Millington D S, Stanley C A, Riely C A, Hyans J S. Am J Gastroenterol. 1996;91:2293–2300. [PubMed] [Google Scholar]
- 28.Battaile K P, McBurney M, Van Veldhoven P P, Vockley J. Biochim Biophys Acta. 1998;1390:2157–2164. doi: 10.1016/s0005-2760(97)00185-9. [DOI] [PubMed] [Google Scholar]
- 29.Eder M, Krautle F, Dong Y, Vock P, Kieweg V, Kim J J, Strauss A W, Ghisla S. Eur J Biochem. 1997;245:600–607. doi: 10.1111/j.1432-1033.1997.00600.x. [DOI] [PubMed] [Google Scholar]
- 30.Bertrand C, Largilliere C, Zabot M T, Mathieu M, Vianey-Saban C. Biochim Biophys Acta. 1993;1180:327–329. doi: 10.1016/0925-4439(93)90058-9. [DOI] [PubMed] [Google Scholar]
- 31.Vianey-Saban C, Divry P, Brivet M, Nada M, Zabot M T, Mathieu M, Roe C. Clin Chim Acta. 1998;269:43–62. doi: 10.1016/s0009-8981(97)00185-x. [DOI] [PubMed] [Google Scholar]
- 32.Litwin S E, Raya T E, Anderson P G, Litwin C M, Bressler R, Goldman S. Circulation. 1991;84:1819–1827. doi: 10.1161/01.cir.84.4.1819. [DOI] [PubMed] [Google Scholar]