Abstract
Insertion sequences (ISs) are among the smallest and simplest autonomous transposable elements. ISfinder (http://www-is.biotoul.fr/) is a dedicated IS database which assigns names to individual ISs to maintain a coherent nomenclature, an IS repository including >3000 individual ISs from both bacteria and archaea and provides a basis for IS classification. Each IS is indexed in ISfinder with various associated pieces of information (the complete nucleotide sequence, the sequence of the ends and target sites, potential open reading frames, strain of origin, distribution in other strains and available bibliography) and classified into a group or family to provide some insight into its phylogeny. ISfinder also includes extensive background information on ISs and transposons in general. Online tools are gradually being added. At present, it is difficult to visualize the global distribution of ISs in a given bacterial genome. Such information would facilitate understanding of the impact of these small transposable elements on shaping their host genome. Here we describe ISbrowser (http://www-genome.biotoul.fr/ISbrowser.php), an extension to the ISfinder platform and a tool which permits visualization of the position, orientation and distribution of complete and partial ISs in individual prokaryotic genomes.
INTRODUCTION
The massive accumulation of sequenced bacterial genomes over the past decade (3600 complete and ongoing archaeal and bacterial genomes and 170 metagenomes) is providing exciting opportunities for understanding genome organization and evolution. Insertion sequences (ISs) play a key role in these processes. At present, there are two major barriers to easily extracting such information. The first is the quality of annotation. There are two important basic objects to annotate: the IS-associated genes (which encode the transposase, the IS-specific enzyme that catalyzes the strand cleavages and transfers necessary for IS movement together with additional regulatory genes) and the physical DNA ends of the IS which are required for activity. Although the full-length transposase genes are generally annotated, they are often identified as ‘integrase/recombinase’ or ‘hypothetical protein’. On the other hand, the DNA features of mobile elements such as ISs are often not annotated or are incorrectly annotated. Moreover, it is even rarer that partial copies of an IS appear in the annotations. Since these represent scars of previous recombination events and can exist in high numbers, they are important in understanding how the host genome was constituted. The second limitation is in the capacity to easily visualize IS locations on a genome. This would facilitate understanding of their impact on the host genome. We have developed and are enriching a dedicated IS database, ISfinder [www-is.biotoul.fr; (1)] which assigns names to individual ISs to maintain a coherent nomenclature; is an IS repository including >3000 individual ISs from both bacteria and archaea; and provides a basis for IS classification. Here we describe ISbrowser (http://www-genome.biotoul.fr/ISbrowser.php), an addition to the ISfinder platform, which has been designed and implemented to address this second limitation and is an important aid in interpretation of the impact of ISs on genome structure and function.
ISbrowser OVERVIEW
ISbrowser has been designed to provide a body of information concerning the IS content of sequenced prokaryotic genomes. It includes only those genomes which have been expertly annotated and verified by ISfinder annotators and is regularly supplemented with additional genomes. Existing genomes will also be regularly updated when new types of IS appear in the ISfinder database. This process will be greatly improved and accelerated in the near future by the addition of a semiautomatic annotation tool, ISsaga (Varani, A. et al., in preparation).
The major feature of ISbrowser is the visualization tool: a circular graphic representation of each genome in which the positions and orientations of ISs and their family attributions are shown. Individual complete and partial ISs are distinguished by a colour code. Additional details concerning a given IS can be obtained simply by clicking on the each individual example. The ISbrowser suite also includes sets of tables which provide a more detailed picture of the IS content and permit the user to: visualize individual multi- or single-copy ISs on the genome; determine the content in user-defined subregions of the genome; obtain alignments of multicopy ISs (the ends—IRs, the entire DNA and amino acid sequences); and obtain information on the IS family through a link to the ISfinder information section. A detailed description of the individual components is provided below as a manual to help potential users.
ISbrowser TOOL
ISbrowser replaces the previous Genome section of ISfinder. It was implemented as a relational database using MySQL (http://www.mysql.com/). CGview (http://wishart.biology.ualberta.ca/cgview/) and Muscle (http://www.drive5.com/muscle/), together with Jalview (2), were used for graphical genome representation and alignment, respectively.
Use of the ISbrowser tool
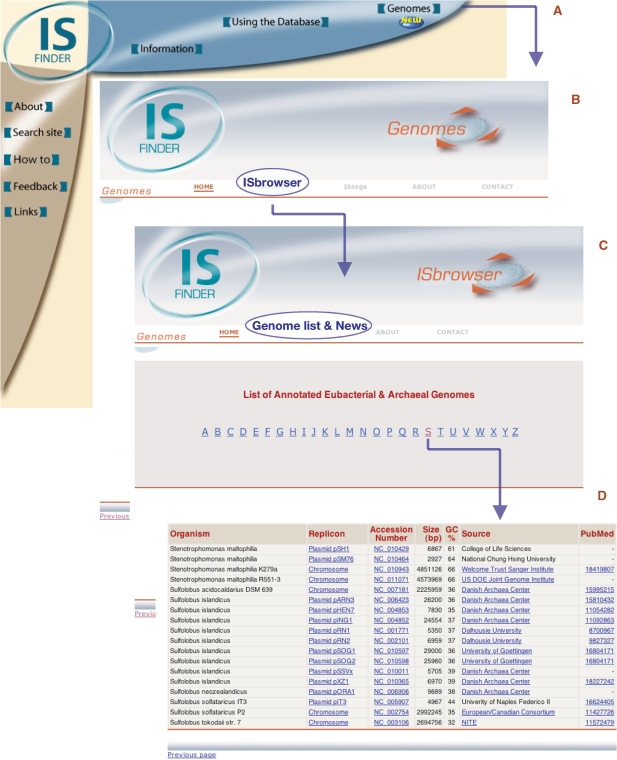
ISbrowser is accessed directly from the ISfinder home page (Figure 1A). The ‘Genomes’ tab gives access to a genome home page (Figure 1B) where the user is presented with a list of ‘tabs’. ‘Home’ (underlined in orange) indicates that the user is on the Genome section home page; ‘ISbrowser’ provides a link toward the browser home page. ‘ISsaga’ is under construction and will provide a link to a pipeline (ISsaga) facilitating rapid semiautomatic IS annotation for outside users and will be described elsewhere (Varani, A. et al., in preparation). ‘About’ provides a concise description of the section content. ‘Contact’ provides relevant addresses and contact information for enquiries.
Figure 1.
The pathway to ISbrowser. (A) The ISfinder home page. (B) The Genome home page. (C) The ISbrowser home page showing the genome list function. (D) The genome list.
ISbrowser home page menu
The ‘ISbrowser’ tab gives access to the ‘ISbrowser home page’ Menu (Figure 1C). The user is presented with a list of ‘tabs’ and, for rapid access, a list of Annotated Eubacterial and Archaeal Genomes in alphabetical order with links to completed prokaryotic genomes annotated and quality controlled by the ISfinder annotators. Choice of a letter generates a complete list of all replicons from organisms whose genera name begins with that letter (Figure 1D).
‘Home’ (underlined in orange) indicates that the user is on the ‘ISbrowser home page’. The entire list of genomes in alphabetical order can be accessed using the ‘Genome List & News/Genome List’ tab.
‘Genome List & News’ has two subsections: ‘News’ that includes updates of the database, new genomes online, new tools, etc. and ‘Genome List’. In turn, ‘Genome List’ has two subsections: ‘Search Genome’, permitting a search for a given genome either using an accession number or the organism name, and ‘Genome List’ that provides a list of all genomes currently in the database. A genome is defined as the genetic material carried by the organism and includes all chromosomes and plasmids. These are entered as separate objects together with their accession number, size in base pairs, average GC%, the source (i.e. the organization responsible for sequencing and/or assembly and overall annotation) and the PubMed link to the original article describing the genome sequence.
‘About’ provides a concise description of the section content and ‘Contact’ provides relevant addresses and contact information for enquiries.
Individual replicon menu
Following a link for a given replicon from the alphabetical list on the ‘ISbrowser home page’ (Figure 1C) or from ‘Genome List’ (Figure 1D) leads to a second page which includes the organism name, genome accession number and taxonomy together with statistics on the number of full-length and partial ISs in a given annotated genome, and the number of IS-related base pairs proportion of IS DNA contained by the replicon. The user also has access to other replicons from the same or related organisms from the same genera. The user is presented with a list of ‘tabs’. ‘Home’ returns the user to the home page of ISbrowser.
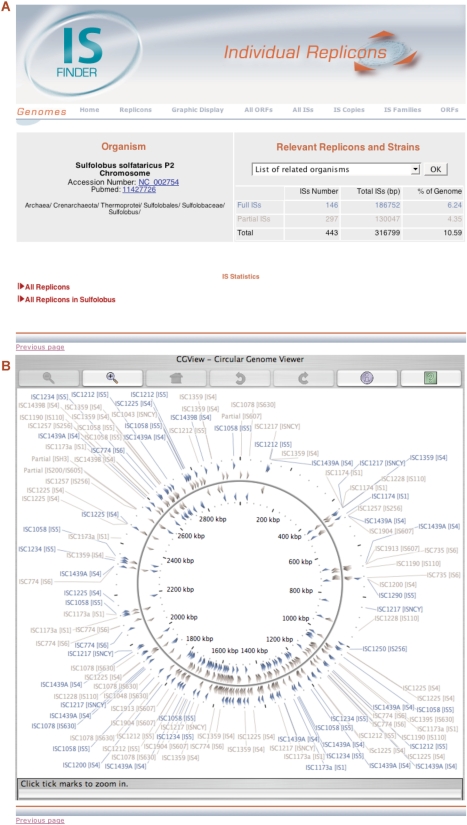
‘Replicons’ (Figure 2A) provides access to ‘IS statistics’ with two tabs, ‘All replicons in a given species’ and ‘All replicons in a given genus’; ‘Graphic Display’ (Figure 2B) tool uses CGview (3) and shows the position and orientation of both full-length and partial IS copies (indicated by their colour) on a circular genome map. This contains zoom and navigation functions. Each IS included in the graphic display tool is labelled by its official name. Its family/subgroup allocation (if any) is indicated in square brackets and is linked to a database entry (Figure 3, ‘Individual IS file’ section).
Figure 2.
The Individual Replicon Page using Sulfolobus solfataricus P2 chromosome as an example. (A) The Individual Replicon Entry Page showing IS statistics. (B) The Graphic Display page showing the S. solfataricus P2 chromosome.
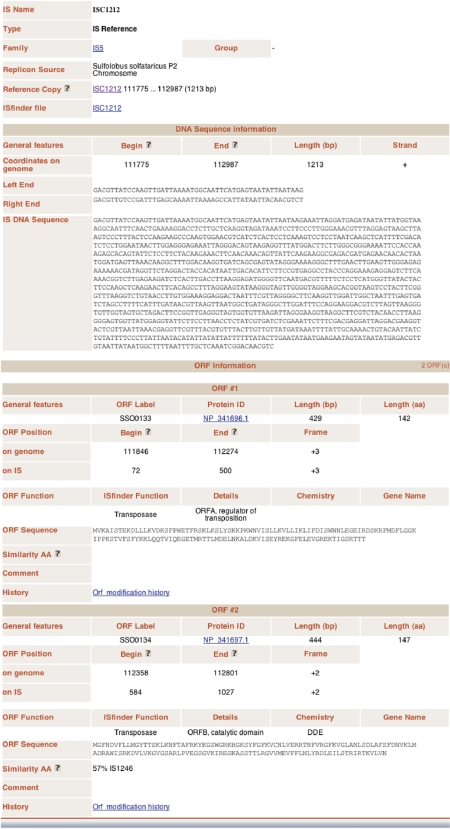
Figure 3.
The individual IS file.
‘All Orfs’ generates a table containing ‘IS Name’ (with a link to the reference IS copy database entry), ‘Orf Name’ (with a link to the Uniprot entry), ‘Family’ (IS family), ‘Orf Begin’ (expressed as genome coordinates), ‘Orf End’ (expressed as genome coordinates), ‘Strand’ (showing orientation), ‘Length’ (in base pairs and amino acids) and ‘ORF Function’ (this provides a description of the functions of all genes included in the IS).
‘All ISs’ is structured similarly to ‘All Orfs’. It generates a table containing the DNA annotations: ‘IS Name’ (note that ISfinder does not generally assign names to partial IS copies but those which do carry a name have been published as such by the initial investigators); ‘IS Family’ (ISfinder defined); ‘IS Family group’ (the subgroup within the family to which the IS belongs, when applicable); ‘Strand’ (indicating the orientation in the genome); ‘Full IS coordinates’ (genome coordinates for the ‘begin’, left end and ‘end’, right end); ‘Length’ (IS length in base pairs); ‘Partial IS coordinates’ (genome coordinates for the beginning and end of partial IS copies); ‘Partial IS coordinates on IS’ (the part of the entire reference IS covered by the partial IS); ‘Comments’.
‘IS Copies’ generates a table with the number of full and partial copies of each IS. This tab provides a form allowing sorting by specific IS names. Choosing a single or any combination of ISs on the right scrolling list and pressing the ‘submit’ button displays the distribution of members of a single or multiple ISs. The results of this query are presented by tables accessible via four tabs: ‘ORF’ displays an identical table to that of ‘All ORFS’ but for a chosen IS. ‘IS List’ displays an identical table to that of ‘All ISs’ but for a chosen IS. ‘CGview Map’ displays an identical figure to that of ‘Graphic Display’ but for a chosen IS. ‘Alignment’ gives access to Jalview applets for alignment of full-length and ‘partial’ DNA as well as ‘IRL’ and ‘IRR’. The tool also permits the user to define a given region of interest in the genome.
‘IS Families’ generates a table with the number of full and partial copies of each IS family. This tab provides a form allowing sorting by specific IS families. Choosing a single or any combination of IS families on the right scrolling list and pressing the ‘submit’ button displays the distribution of members of a single or multiple ISs. The results of this query are presented by tables accessible via three tabs, ‘ORF’, ‘IS List’ and ‘CGview Map’, which provide similar information to those given in ‘IS Copies’.
‘ORFS’ allows the user to define a subsection of the genome (left) and to view its IS content or to search for a given IS-associated orf (right).
The individual IS file
The individual IS file (Figure 3) can be accessed from individual ISs displayed in CGview (Figure 2B) or from each citation of the IS from tabs ‘all orfs’ and ‘all ISs’. This includes the following information: ‘IS name’; ‘Type’ (whether the IS is a Reference, Full or Partial IS copy;); ‘Family’ and ‘Group’ (if appropriate); ‘Replicon Source’ (name of the replicon in which it occurs); ‘Reference Copy’ (for an IS which is present in >1 copy in each genome, a single given copy of the IS is defined as the reference copy); ‘ISfinder file’ (link to the database entry for the IS in ISfinder).
‘DNA sequence information, includes’: ‘General features’ (‘Begin’ identifies the first nucleotide of the left end of the IS which is closest to the transposase promoter, ‘End’ identifies the last nucleotide of the right end, ‘Length’ gives the overall IS length, ‘Strand’ defines the orientation of the IS, ‘Left End’ and ‘Right End’ indicate the first and last 50 bp); ‘DNA Sequence’ gives the entire nucleotide sequence; ‘Similarity DNA’ indicates the percentage identity with the reference copy.
‘Orf Information’ includes the number of orfs carried by the IS. Information for each orf includes: General features (‘ORF label’ in the annotated genome, ‘Protein ID’ is the link to Uniprot, ‘Length’ in base pairs, ‘Length’ in amino acids); ‘ORF Position’ on the genome with genome coordinates and on the IS (‘Begin’ indicates the first nucleotide of the start codon, ‘End’ indicates the last nucleotide of the stop codon, ‘Frame’ gives the relative reading frame); ‘ORF Function’ (‘ISfinder function’ defines whether the gene is the transposase or an accessory gene, ‘Details’ defines the precise gene function, ‘Chemistry’ defines the transposase catalytic chemistry, ‘Gene Name’ gives the accepted genetic nomenclature); ‘ORF Sequence’ presents the predicted amino acid sequence; ‘Similarity aa’ gives percent similarity with the reference copy or the closest relative in the ISfinder database.
CONCLUDING REMARKS
ISfinder has been operational for several years and we expect an increasing number of online submissions both from individuals (an aspect which at present functions relatively well) and especially from the genome sequencing projects (which at present involves only a limited number of sequencing centers).
One general goal of ISfinder will be to interact with other complementary specialized databases such as those including bacteriophages, plasmids, integrons, recombinases and genomic islands. Future work will involve addition of such specialized databases to the ISfinder suite and extension to eukaryotic mobile genetic elements. One ongoing project is to provide an interface with ACLAME (A CLAssification of genetic Mobile Elements: http://aclame.ulb.ac.be/) (4).
Finally, ISfinder functions as a research tool. Thorough and systematic bacterial genome analysis regularly identifies new phylogenetically related groups and families and this aspect of the database will undoubtedly continue to provide information on the influence of ISs on genome structure, their distribution between genera and species and their degree of spread within and between ecological niches.
FUNDING
Funding for open access charge: Centre National de la Recherche Scientique (CNRS), France.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
This site was designed by David Villa (IBCG, Toulouse) and is administered, maintained and upgraded by Jocelyne Pérochon (LMGM) and Sylvain Corcoral (LMGM). It is curated by Patricia Siguier and Mick Chandler (LMGM, Toulouse). Further information concerning technical aspects of this site can be obtained from P. Siguier, J. Pérochon or M. Chandler. ISfinder is supported by the CNRS (France). We would like to acknowledge the help of Yves Quentin, Edith Gourbeyre, Gwennaele Fichant and Alessandro Varani for additional help and members of the Mobile Genetic Element group (Adeline Achard, Guy Duval-Valentin, Laure Lavatine, Brigitte Marty, Bao Ton-Hoang and Oliver Walisko) for reading the manuscript and for discussions.
REFERENCES
- 1.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 4.Leplae R, Hebrant A, Wodak SJ, Toussaint A. ACLAME: A CLAssification of Mobile genetic Elements. Nucleic Acids Res. 2004;32:D45–D49. doi: 10.1093/nar/gkh084. [DOI] [PMC free article] [PubMed] [Google Scholar]