Abstract
PROSITE consists of documentation entries describing protein domains, families and functional sites, as well as associated patterns and profiles to identify them. It is complemented by ProRule, a collection of rules based on profiles and patterns, which increases the discriminatory power of these profiles and patterns by providing additional information about functionally and/or structurally critical amino acids. PROSITE is largely used for the annotation of domain features of UniProtKB/Swiss-Prot entries. Among the 983 (DNA-binding) domains, repeats and zinc fingers present in Swiss-Prot (release 57.8 of 22 September 2009), 696 (∼70%) are annotated with PROSITE descriptors using information from ProRule. In order to allow better functional characterization of domains, PROSITE developments focus on subfamily specific profiles and a new profile building method giving more weight to functionally important residues. Here, we describe AMSA, an annotated multiple sequence alignment format used to build a new generation of generalized profiles, the migration of ScanProsite to Vital-IT, a cluster of 633 CPUs, and the adoption of the Distributed Annotation System (DAS) to facilitate PROSITE data integration and interchange with other sources. The latest version of PROSITE (release 20.54, of 22 September 2009) contains 1308 patterns, 863 profiles and 869 ProRules. PROSITE is accessible at: http://www.expasy.org/prosite/.
GENERALITIES
The PROSITE database uses two kinds of signatures or descriptors to identify conserved regions, i.e. patterns and generalized profiles, both having their own strengths and weaknesses defining their area of optimum application. Each PROSITE signature is linked to an annotation document where the user can find information on the protein family or domain detected by the signature, such as the origin of its name, taxonomic occurrence, domain architecture, function, 3D structure, main characteristics of the sequence, domain size and literature references (1). PROSITE signatures are also associated with corresponding ProRules (2), which contain information for the automated annotation of domains in the UniProtKB/Swiss-Prot database. Some of the information stored in ProRule can be accessed via ScanProsite, which provides additional features such as active sites or disulfide bonds associated with a particular domain (3).
Since the latest NAR database issue paper (4), PROSITE has increased its number of signatures to 1559 documentation entries, 1308 patterns, 863 profiles and 869 ProRules. Most of the newly integrated profiles were constructed with a new method, apsimake, which makes use of an annotated multiple sequence alignment (AMSA) and will be described elsewhere. The number of patterns has decreased since some patterns had too many false positive matches and have been replaced by a profile covering the same domain. The list of deleted PROSITE signatures as well as the ones which replace them is available in the psdelac.txt file.
While other protein domain databases such as Pfam (5) aim to be comprehensive and to a maximum sequence coverage, PROSITE concentrates on precise functional characterization, which can be used for protein database annotation. These efforts are time consuming, which is reflected by a reduced number of protein domains as compared with Pfam. Some PROSITE profiles might also be less sensitive as they are intended to cover domains over their entire length with the best possible alignment. Partial matches are strongly penalized, which allows full length detection of domain but decreases the sensitivity. Truncated domains due to mispredicted protein sequences might be missed.
In collaboration with PeroxiBase (6), PROSITE has developed a strategy to construct profiles specific for the different subfamilies of the peroxidase family. This strategy has been improved and applied to other complex families such as the small GTPases. This approach will be pursued in the future to provide the scientific community with a large number of subfamily profiles for more accurate function inference. More specific profiles for subfamilies allow better functional prediction, as different subfamilies can have various, although related, functions.
PROSITE is extensively used by UniProtKB/Swiss-Prot curators to annotate domains with the help of ProRule, which provides functional annotation associated with a specific domain in a UniProtKB/Swiss-Prot format (2). Among the 983 different types of (DNA-binding) domain, repeat or zinc finger found in UniProtKB/Swiss-Prot, 696 (∼70%) are annotated with the help of PROSITE descriptors using information from ProRule. The usage of PROSITE and ProRule during the annotation process facilitates the transfer of the positions of biologically meaningful sites, such as active or binding sites and disulfide bonds, and ensures that all useful information that can be associated with a domain will be added to UniProtKB/Swiss-Prot entries. This is a guarantee for homogeneous domain annotation.
A NEW CLUSTER FOR IMPROVED RAPIDITY
ScanProsite—http://www.expasy.org/tools/scanprosite/—is a web-based tool for detecting PROSITE signature matches in protein sequences (3). For many PROSITE profiles, the tool makes use of ProRules to detect functional and structural intra-domain residues. The increase of data in UniProtKB and the analyses of subfamilies by searching these data with several profiles simultaneously have incited us to speed up the ScanProsite performance. To do so, ScanProsite was migrated to the Vital-IT Center for high-performance computing (cluster of 633 CPUs) of the Swiss Institute of Bioinformatics (7). To increase the reliability of ScanProsite, the old infrastructure has been kept as a fallback in case of failure of the Vital-IT cluster.
With the advent of high-throughput sequencing, users need to analyze large sets of proteins. To respond to these requests, we make use of Vital-IT to allow PROSITE users to submit large sets of proteins to ScanProsite. We are currently building the tools and procedures that will allow the analysis of full proteomes with PROSITE.
Additionally to the use of a new cluster, we want to improve the speed of ScanProsite by using a heuristic approach to perform a fast prescan to detect protein sequences to which the usual scan should be applied. This way, we might get the usual alignment necessary to apply ProRule to detect functional residues. We are currently evaluating different heuristic methods such as BLAST (8) or HMMER3 (9). Preliminary results show a speed increase of 15–20 times.
AMSA TO BUILD GENERALIZED PROFILES
Manual modifications of generalized profiles are normally performed within PROSITE to improve the models generated by the PFTOOLS software (1). This manual editing is an important added value of the PROSITE database. However, these adjustments are understandable only to experts with good knowledge of the generalized profile syntax, and tracking of modifications by different users can be difficult. Ideally, the model adjustment should be done directly in the input multiple sequence alignment (MSA) in the form of annotation. Additionally, there is an increasing need to produce profile models not only to detect distant homologs but also to classify subfamily sequences and to transfer annotation at the residue level automatically. Thus we need profile models tuned for classification and for high quality alignments. To achieve this, we need to explicitly change parameters used for the construction of the profile in a position-dependent manner, e.g. change the substitution matrix used for pseudocounts in one region of the alignment, give different weight to the pseudocounts with respect to the observed residues, change gap penalties, etc. The position specific parameters should be explicitly included in the MSA used to build the profile.
To fulfill all our requirements, we have developed a strategy to clearly distinguish between the AMSA, containing all the information needed to build the profile, and the final numerical prediction model, usually a scoring matrix (generalized profiles in our case).
We have defined a data structure to store AMSA. The grammar and recommendations of AMSA (v1.0) are given in Figure 1. The AMSA format is organized as a standard MSA file. Each sequence entry is represented in the standard FASTA format. Sequences contain symbols from the residue alphabet and the gap symbols. Annotation is added as standard FASTA-like sequences that we refer to as annotation layers, with symbols from an ad hoc alphabet. The identifier of annotation layers should start with the symbol ‘#’ and in the description field of the FASTA format each symbol of the annotation alphabet is paired to its value (symbol ∼ value) (see Figures 1 and 2). We opted for this representation and not the Stockholm format (7) because it is compact and any pair symbol ∼ value (except for a few symbols, see Figure 1A legend) can be defined in the description field and any annotation layer can be added without restrictions. The AMSA format is also simple to parse by standard programs and easy to edit by hand or using the Jalview MSA editor, which in its latest version supports the AMSA syntax (see Figure 2B) (8,9).
Figure 1.
(A) AMSA 1.0 grammar in Extended Backus-Naur Form. Explanation of the grammar symbols: ‘=’ assign operator; ‘()’ grouping operator; ‘[]’ optional (0 or 1 occurrence); ‘{}’ repetition (0 or more occurrences); ‘-’ exclude following symbol; ‘|’ alternation (or), ‘??’ special sequence. Note that any visible symbol is accepted for sequence and annotation alphabets, except the special symbols ‘#’, ‘=’, ‘∼’ and ‘>’. (B) Recommendations for the AMSA 1.0 format. An AMSA file consists of aligned sequences and annotation represented in FASTA format. Is possible to precede the sequence and annotation block with a version header followed by general comments (lines starting with '#') and global annotation (lines starting with ‘##’ followed by a key = text); in this case, we must terminate the sequence and annotation block using symbols ‘#//’. Any ad hoc alphabet can be used to describe sequences and annotation. The meaning of the alphabet used by an annotation layer can be explicitly represented as pairs value ∼ symbol in its description field. Sequence- (or annotation)-specific annotation is added after the ID field using key = value (or key = ‘text with white-spaces’). Sequence residues are annotated using a cross-reference to an annotation layer (key = # residue_annotation_ID). Though not required by the specification, we suggest to include the reference sequence ID in the residue_annotation_ID to facilitate the visual mapping from annotation to sequence. Columns of the MSA are annotated using annotation layers not linked to one or multiple sequences (see Figure 2 for an example). Two keywords are reserved in the current format: ‘DEFAULT’ is used to set a default value for the ‘-’ symbol of the annotation sequence; ‘SYM_LEN’ defines the number of characters required to encode a symbol. The SYM_LEN = digits permits to extend the format of sequences and annotations to any alphabet, e.g. codons (SYM_LEN = 3), to store PDB coordinates (SYM_LEN = 8 to store up to 6 digits, dot and sign), etc. If SYM_LEN is not specified, its default value should be 1 (one character per symbol). Note that we can achieve the same result by encoding multi-character symbols on a corresponding number of layers, this having the advantage that standard MSA editors can accept the format.
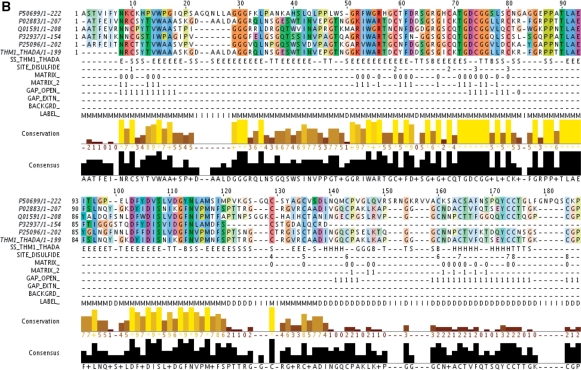
Figure 2.
(A) Example of an AMSA. Protein sequences containing symbols from the amino acid alphabet and ‘-’ to represent gaps are represented in the standard MSA format at the beginning of the file. Annotation layers (layers beginning with ‘#’) are in the second part of the AMSA file and they contain any ad hoc alphabet. The meaning of each symbol of the alphabet is described by the symbol ∼ value pairs in the description field of each layer. Note that the symbol ‘-’ in annotation denotes a default value described by the DEFAULT key. Annotation attached to the individual sequences is represented as key = value pairs (e.g. weight of the sequence). Annotation attached to individual residues of a sequence is described in a cross-referenced layer: in the example the ‘ss_layer’ cross-reference of THM1_THADA points to the secondary structure of the protein in layer #_SS_THM1_THADA. MSA column annotation is used to label disulfide bridges (#_SITE_DISULFIDE) where numbers represent the cysteine couple and values represent the expected symbol (C for cysteine in this example). The remaining layers represent parameters of the profile: the topology of the model (#_LABEL_), the two matrices used to generate pseudocounts (layers #_MATRIX_ and #_MATRIX_2) with the respective position-specific weights of the pseudocounts. (B) The same alignment viewed in Jalview.
Annotation can be attached to the different dimensions of the AMSA. (i) per-sequence: e.g. sequence weight, cross-references, etc. using the pair key = value, e.g. weight = 0.2. (ii) per-column: features are stored in an annotation layer where each position contains a symbol associated with a value as defined in the description field. This annotation refers to biological information, such as active sites or post-transcriptional modification, or information used for the construction of the profile model, as the scoring system associated with each column. (iii) per-residue: annotation layers can be associated with a single sequence using a cross-reference; each position contains a symbol associated with annotation for the corresponding residue in the sequence, e.g. the secondary structure of the sequence. (iv) global: global alignment annotation can be added at the beginning of the AMSA using the characters “##” followed by the pair key = text, e.g. to incorporate phylogenetic information. Lines starting with a single symbol ‘#’ are considered as free text comment lines (see Figure 1 for the full specifications). Note that the AMSA v1.0 specifications permit to convert the interleaved Stockholm format to the noninterleaved AMSA format without any loss of information by using the same keywords.
We have developed msa2amsa, a program to add annotation to an MSA file to build an AMSA containing all information required for the construction of the final profile scoring matrix. The program parameters can be adjusted to produce AMSA annotation to build profiles for distant homologous sequence discovery, subfamily classification and to produce alignments for automatic annotation. A second program, apsimake, reads an AMSA file and produces the final scoring matrices (generalized profiles in PROSITE). The profiles constructed using this new method generally perform better than linear profiles built using such classical methods as PFTOOLS and HMMER (results will be published in a separate article).
Within PROSITE, we now use AMSA to store MSAs combined with annotation and msa2amsa/apsimake to build generalized profiles fine-tuned for classification and automatic residue annotation.
PROSITE DISTRIBUTED ANNOTATION SYSTEM SERVICES
In order to facilitate data integration by the InterPro consortium and data exchange between members of the consortium (13), PROSITE has adopted the Distributed Annotation System (DAS) (14). DAS is a lightweight decentralized system for exchanging and aggregating data from a number of heterogenous databases using a common biological data exchange standard (DAS XML specification). DAS has become the standard programmatic exchange protocol for biological data annotation.
We created a PROSITE DAS annotation server providing features (PROSITE matches on a specific UniProtKB entry), and MSAs (alignment of match regions on UniProtKB entries for a specific PROSITE motif) using the ProServer (http://www.sanger.ac.uk/Software/analysis/proserver/) Perl framework (15).
In order for our service to be easily discoverable, we registered it on the central DAS registry (http://www.dasregistry.org) (16). It can be monitored through http://www.dasregistry.org/listServices.jsp?keyword=prosite&cmd=keyword
The implemented DAS commands are:
(i) for MSAs:
- alignment (e.g. http://proserver.vital-it.ch/das/prositealign/alignment?query=PS50808);
(ii) for multiple sequence features:
- features (e.g. http://proserver.vital-it.ch/das/prositefeature/features?segment=P08487).
PROSITE matches, including low confidence ones (not shown in InterPro), within a specific UniProtKB entry (optional: start, end of a range within the protein sequence);
- types (e.g. http://proserver.vital-it.ch/das/prositefeature/types);
- sequence (e.g. http://proserver.vital-it.ch/das/prositefeature/sequence?segment=P08487).
In addition to its use within the InterPro consortium, the PROSITE DAS service can be accessed by other users who want to make use of PROSITE data and integrate them (with other sources) with their own personal data in a convenient and standardized way as several independent annotation servers can be connected to a reference sequence. The Dasty viewer (http://www.ebi.ac.uk/dasty/) is a popular proteins feature viewer (17) that uses the DAS protocol and which can be accessed from UniProtKB entries (third-party data).
For example, with http://www.ebi.ac.uk/dasty/client/ebi.php?q=P08487, you can see PROSITE ‘polypeptide_domain’ and ‘polypeptide_repeat’ (including low confidence matches) features on UniProtKB protein P08487, aggregated with data from other sources.
The database behind the PROSITE DAS service is updated synchronously with PROSITE releases; therefore, the DAS service will stay up-to-date.
FUNDING
FNS project grant (315230-116864) and European Union grant (213037). PROSITE activities were also supported by the Swiss Federal Government through the Federal Office of Education and Science. Funding for open access charge: FNS Project grant (315230-116864).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Marco Pagni for helpful discussions and ideas and Andrea H. Auchincloss for critical reading of the manuscript.
REFERENCES
- 1.Sigrist CJA, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, Bairoch A, Bucher P. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinformatics. 2002;3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 2.Sigrist CJA, de Castro E, Langendijk-Genevaux PS, Le Saux V, Bairoch A, Hulo N. ProRule: a new database containing functional and structural information on PROSITE profiles. Bioinformatics. 2005;21:4060–4066. doi: 10.1093/bioinformatics/bti614. [DOI] [PubMed] [Google Scholar]
- 3.de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJA. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koua D, Cerutti L, Falquet L, Sigrist CJA, Theiler G, Hulo N, Dunand C. PeroxiBase: a database with new tools for peroxidase family classification. Nucleic Acids Res. 2009;37:D261–D266. doi: 10.1093/nar/gkn680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vital-IT. Available at: http://www.vital-it.ch/ [Accessed August 27, 2009] [Google Scholar]
- 8.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.HMMER. Available at: http://hmmer.org/ [Accessed September 28, 2009] [Google Scholar]
- 10.Stockholm format. Available at: http://sonnhammer.sbc.su.se/Stockholm.html [Accessed August 18, 2009] [Google Scholar]
- 11.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 12.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn RD, Stalker JW, Jackson DK, Kulesha E, Clements J, Pettett R. ProServer: a simple, extensible Perl DAS server. Bioinformatics. 2007;23:1568–1570. doi: 10.1093/bioinformatics/btl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prlić A, Down TA, Kulesha E, Finn RD, Kähäri A, Hubbard TJP. Integrating sequence and structural biology with DAS. BMC Bioinformatics. 2007;8:333. doi: 10.1186/1471-2105-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimenez RC, Quinn AF, Garcia A, Labarga A, O’Neill K, Martinez F, Salazar GA, Hermjakob H. Dasty2, an Ajax protein DAS client. Bioinformatics. 2008;24:2119–2121. doi: 10.1093/bioinformatics/btn387. [DOI] [PubMed] [Google Scholar]