Abstract
Chemical Entities of Biological Interest (ChEBI) is a freely available dictionary of molecular entities focused on ‘small’ chemical compounds. The molecular entities in question are either natural products or synthetic products used to intervene in the processes of living organisms. Genome-encoded macromolecules (nucleic acids, proteins and peptides derived from proteins by cleavage) are not as a rule included in ChEBI. In addition to molecular entities, ChEBI contains groups (parts of molecular entities) and classes of entities. ChEBI includes an ontological classification, whereby the relationships between molecular entities or classes of entities and their parents and/or children are specified. ChEBI is available online at http://www.ebi.ac.uk/chebi/. This article reports on new features in ChEBI since the last NAR report in 2007, including substructure and similarity searching, a submission tool for authoring of ChEBI datasets by the community and a 30-fold increase in the number of chemical structures stored in ChEBI.
INTRODUCTION
Recent efforts to understand the metabolism of organisms on a systematic level have created a demand for data on small organic molecules and their reactions, traditionally a chemical domain. The data of interest include the chemical structures themselves and their spectroscopic data, as well as kinetic and thermodynamic parameters of chemical reactions. These data are typically grouped around small molecule structures which again can be marked up as substrates of enzymes, as participants in a particular metabolic pathway or as subjects of transport by a membrane protein. Wherever molecular biology resources include such data on small molecules, there is a need for a ‘unified resource of curated small molecule structures’ to point to, a ‘dictionary of chemical compounds'. Furthermore, there is a need for an ‘ontology', classifying chemical compounds according to their biological role, chemical nature, etc.
This demand has led to the establishment of a ‘database’, ‘dictionary’ and ‘ontology’ of Chemical Entities of Biological Interest (ChEBI) at the European Bioinformatics Institute (EBI) (1). ChEBI datasets are molecule-centric with a number of annotations grouped around the 2D molecular graphs (connection tables) of small molecules. Each entry is manually annotated by expert annotators before being released. As a dictionary, the nomenclature provided includes an unambiguous ChEBI recommended name, IUPAC names, International Nonproprietary Names (INNs) and synonyms, all manually annotated. Where feasible a molecular graph is provided accompanied by the chemical structural representations InChI, InChIKey and SMILES. Additional chemical data such as formula, mass and charge are provided. Each entry is then extensively cross-referenced. External databases link to ChEBI via the unique and stable ChEBI identifier.
We have reported on ChEBI in an earlier, first article about ChEBI in the 2007 NAR Database issue and we will restrict ourselves here to describing the novelties introduced into ChEBI since then, which include a submission tool, news in the ontology development, substructure and similarity searching, links to patents and a 30-fold increase in the number of structures provided.
NEW FEATURES SINCE 2007
Many, many more compounds
In 2008, the EBI was awarded a substantial grant to support the transfer of a large collection of information on the properties and activities of drugs and a large set of drug-like small molecules from the publicly listed company Galapagos NV into the public domain. This data, now named ChEMBL (http://www.ebi.ac.uk/chembl/), consisted of the protein targets and their associated bioactive small molecules. The small molecule data consisted of a chemical structure and associated synonyms. These were manually annotated from the original publication. The data were loaded into ChEBI and associated properties such as formulae, mass and charge automatically generated from the chemical structures. Duplicate entities were found by using the InChI and were merged into a single ChEBI entity.
Although the ChEMBL data had been manually annotated, some core information that is normally part of the ChEBI annotation process was missing. Examples of these are manually annotated IUPAC names and additional cross-references to core resources such as KEGG COMPOUND. In order to distinguish between fully annotated ChEBI entities and partially annotated ChEMBL data we devised a starring system in which fully annotated ChEBI entities are allocated three stars and partially annotated ChEMBL entities are allocated two. These are clearly indicated on the entry pages on the ChEBI public website. The ChEMBL data are regularly updated within ChEBI.
The ChEMBL data are distributed along the normal channels available to other datasets in ChEBI through the ChEBI website. The data can also be queried programmatically via the ChEBI Web Service. The full dataset is downloadable via the ChEBI download formats such as tab delimited flat files, database dumps and MDL SD file formats. Furthermore, ChEBI has introduced a new MDL SD file export to distribute its chemical structures. For users just wishing to retrieve the chemical structure, name and identifier they can use the smaller file, namely ChEBI_lite.sdf, while for those users wanting the entire dataset then ChEBI_complete.sdf can be used. Note that ChEBI_complete.sdf does not extract any ontology relationships but merely the data record for each entry. All data formats allow download of only three star entries as well as downloads of both two and three star entries.
In addition to the ChEMBL data, ChEBI was also populated with data from the PDBeChem database. PDBeChem (2) is a consistent and enriched library of ligands, small molecules and monomers that are referenced as residues and hetgroups in PDB entries. The PDBeChem data are loaded into ChEBI on a regular basis. Data which are duplicated are merged into existing ChEBI entities and additional data items are annotated to ChEBI standards. An example of such an entry can be viewed for FAD (http://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:16238). The entry is linked back to PDBeChem via the three letter code.
The ChEBI database now contains >440 000 entities as illustrated in Table 1.
Table 1.
ChEBI entry statistics illustrating the 30-fold increase in data
| Number of entities in ChEBI, July 2009 | 18 414 |
| Number of unique entities loaded from ChEMBL | 437 765 |
| Number of entities merged with existing ChEBI annotated entities | 2220 |
| Total entities in ChEBI after loading ChEMBL data | 456 179 |
Submission tool
To alleviate the backlog of user requests for inclusion into ChEBI, and to invite the community to participate more directly in the future growth and development of ChEBI, we have developed a web-based software utility to enable direct user submissions. User submissions are then publicly available (after the next release cycle) and cited to the submitter (although the submitter has the option to remain anonymous if he/she wishes).
The ChEBI submission tool is available online at https://www.ebi.ac.uk/chebi/submissions. It is secured and each user must apply for a login username and password. During this application process, the user's; email address is verified and the user's; credentials are checked by the ChEBI team to ensure that fictitious users are not authorized. Once a user has been authorized, he/she is able to begin creating submissions.
The minimal information which is required for a ChEBI submission is a name, which must be unique within the database, either a text definition or a chemical structure and a primary classification within the ontology. For example, the term ‘insecticide’ might be submitted to the ontology with definition ‘A substance used to destroy pests of the class Insecta.’ and primary classification ‘is a pesticide (CHEBI:25944)'. Of course, we encourage the submission of the most complete dataset possible, and it is possible to add multiple synonyms and database cross-references, as well as to create multiple relationships within the ontology.
The captured submission is automatically validated for uniqueness, both of name and chemical structure (where applicable), and correctness, such as checking that no cycles have inadvertently been created in the ontology graph structure, and that the ontology relationships which have been specified are allowed between entities of the relevant types. Warnings are issued if any of the captured synonyms and database cross-references map to existing synonyms or cross-references in the database. Final submission is not possible until all errors have been resolved. Once the submission has passed the required validations, it may be submitted to the ChEBI database and will receive a unique ChEBI identifier.
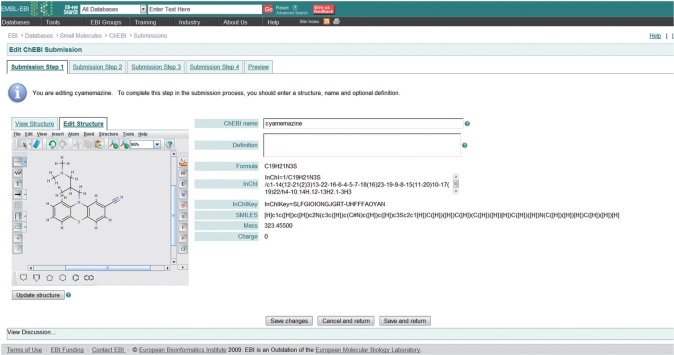
The submitted entry enters the ChEBI database with status ‘SUBMITTED'. With this status, it will become visible to the public as a preliminary entry after the next release cycle (which runs on the last Wednesday of every month). However, at the earliest opportunity the submission will be checked by ChEBI annotators, during which process the data will be enriched with additional synonyms, cross-references and registry numbers, and a chemical structure will be drawn if the submitter did not provide one. Once the annotation process is complete, the entry will transition to status ‘CHECKED’ and will appear as any regular ChEBI entry, although retaining the citation of the original submitter. From time to time it may be found that a submission is inappropriate for ChEBI for some reason, in which case the submitter will be notified with a detailed reason and the submission will transition to status ‘DELETED'. On the other hand if it is found that the same entry already existed within ChEBI but with a different name (so that it was not able to be automatically detected), the submitted entry will be merged with the existing entry and again, the submitter will be notified. Discussions, in the form of electronic comments between the ChEBI annotators and the data submitter, are displayed publicly via the ChEBI web entry page. ChEBI reserves the right to alter any of the submitted data. The submission tool is illustrated in Figure 1.
Figure 1.
Illustrates the first step in the ChEBI Submission procedure allowing the user to input the chemical structure, name and definition.
Ontology development
Changes to the ChEBI ontology have been implemented to address user concerns regarding ambiguous and incorrect usage of the relationships ‘is a’ and ‘is part of'. The parthood relationship ‘is part of’ was formerly used ambiguously to mean several different things, including not differentiating between possible and necessary parthood (3). This has been resolved by the introduction of the relationship ‘has part’ which is the inverse of the ‘is part of’ relationship, replacing all former ambiguous usage of ‘is part of’ with a consistent usage according to the definition of ‘has part’ in the Relationship Ontology (4), namely that if Entity A has part Entity B, then ‘for all’ instances of Entity A, ‘some’ instance of Entity B forms a part of it. This change has allowed for more accurate inference based on assertions within the ontology.
Furthermore, the relationship ‘is a’ was being overloaded within the ontology, being used both to link molecular entities with chemical classes, and to specify the ‘roles’ that chemical entities might enact in various contexts. To resolve this overloading, a new relationship ‘has role’ was introduced and used to link molecular entities to the roles that they enact. Alongside this change, the ontology root terms ‘biological role’ (CHEBI:24432) and ‘application’ (CHEBI:33232) were assigned to a new, broader root term ‘role’ (CHEBI:50906). The domain of the ‘has role’ relationship is the molecular entities within the molecular structure ontology, and the range is the role ontology. Thus, if Entity A has role Role B, then all instances of type Entity A ‘may realise’ the role Role B in an appropriate context. For example, the molecular entity acetylsalicylic acid (CHEBI:15365) has role non-narcotic analgesic (CHEBI:35481), which means that molecules of acetylsalicylic acid may realise the role of being a non-narcotic analgesic under appropriate circumstances (such as when imbibed by a human with a headache).
The introduction of a single role ontology root has allowed the extension of the role ontology to include also chemical role (CHEBI:51086), under which several chemical roles are classified such as solvent (CHEBI:46787) and photochemical (CHEBI:52215). The molecular structure ontology and the role ontology are now disjoint, facilitating the use of automatic reasoners in determining the ancestry of particular molecular entities or roles within the ontology.
After extensive manual annotation, the ‘unclassifieds’ root term has been obsoleted within the ontology and all chemical entities that were linked to it have now been classified within the main ontology.
New search facilities
As the data in ChEBI have grown extensively over the last year a new search interface was introduced. The search interface contains two parts, a text search as well as a structure search. The text search was implemented with Lucene, a full-featured text search engine library written entirely in Java. Lucene provides the standard features of wildcards (*) and fuzzy searches (∼) as well as more advanced features such as range searches and logical operators. Most importantly Lucene is an open source project, thus further underlying ChEBI's; commitment to provide an open source framework for chemistry. The ChEBI text search allows users to search all the data or to filter a search by the categories of ChEBI identifiers, names, database links, formula, SMILES and InChI. Mass and charge can be searched within ranges, for example, one can search for all entities with a mass of between 150 and 300 atomic mass units. Furthermore, searches can be filtered by database; for example, one can search for entities used in the NMRShiftDB (5) or PubChem (6) databases.
The ability to filter on ChEBI ontology terms has also been introduced. This functionality allows one to retrieve all the children of a specific entity based on the relationship given. For example, all cofactors (CHEBI:23357) can be retrieved by entering the term ‘cofactors’ using the ‘has role’ relationship and this will retrieve not only its direct children such as pantothenic acids (CHEBI:25848) but also further entities in the graph related via an ‘is a’ relationship such as NADPH (CHEBI:16474). It also allows retrieval of only those entities with chemical structures by ticking a specific checkbox.
The ChEBI database has introduced chemical structure searching using a new chemical structure search algorithm developed in the open source OrChem (7) project. OrChem is an Oracle chemistry plug-in using the Chemistry Development Kit (CDK) (8). OrChem allows one to perform substructure and similarity searching on an Oracle 11g database. It loads the CDK Java library into Oracle and harnesses the Oracle VM just-in-time (JIT) compiler for much faster execution because it manages the invalidation, recompilation and storage of code without an external mechanism. This has greatly improved the speed of chemical structure searching in ChEBI with the new ChEMBL data. OrChem allows the user to search on groups as well as residues. ChEBI has also implemented an identity search using the InChI as the primary chemical structure identifier.
All the above searches can be combined by using the logical operators AND, OR and BUT NOT. All search results can be exported in MDL SD file, tab delimited or XML format.
Patents and cross-references
In order to provide a portal for use by both the bioinformatics and chemoinformatics communities, ChEBI has extended its cross-linking to other databases. We have used the InChI and InChIKey in identifying chemical compounds from the NMRShiftDB database, which contains organic structures and their nuclear magnetic resonance (nmr) spectra. Furthermore we have used the InChIKey to identify compounds in BRENDA (9), the enzyme information system. We have also linked to IntEnz (10), the integrated relational enzyme database, via its use of cofactors and reaction participants as ChEBI entities. As an extension of these links to IntEnz we also link to Rhea, the biochemical reactions database hosted at the EBI. We have also extended our automatic cross-reference links to include annotated ChEBI identifiers within the ArrayExpress (11), BioModels (12), Reactome (13) and SABIO-RK (14) databases.
ChEBI also contains links to Patent documents which were implemented as a collaboration with the European Patent Office using the Oscar3 toolkit (15). The patents referenced consist of only a subsection of the patents available and are relevant only to the life sciences. The links can be searched using the ChEBI standard search and searches can be filtered to include only compounds referenced in patents (or indeed any of the databases cross-referenced in ChEBI).
Additional data items
To provide a consistent chemical information system for the annotation of biochemical reactions, further chemical properties were added to ChEBI. In addition to the already existing formula, charge and mass were added. Relative molecular, atomic and ionic masses are shown for molecular, atomic and ionic entities, respectively. The relative masses are calculated from tables of relative atomic masses (atomic weights) published by IUPAC and are automatically generated using ChemAxon’s MarvinBeans software. For ions the magnitude of the charge is given in arabic numerals preceded by the sign of the charge. For neutral molecules the charge is indicated as a numerical zero. For instance, the charge of 5,10,15,20-tetrakis(1-methylpyridinium-4-yl)porphyrin (CHEBI:37447) is +4; the charge of borate (CHEBI:22908) is –3.
To aid the identification of molecules via search engines such as Google, an extention of the InChI (16), namely the InChIKey, was introduced. The InChIKey is a 25-character hashed version of the full InChI, designed to allow for easy web searches of chemical compounds. InChIKeys consist of 14 characters resulting from a hash of the connectivity information of the InChI, followed by a hyphen, followed by eight characters resulting from a hash of the remaining layers of the InChI, followed by a single character indicating the version of InChI used, followed by a single checksum character. There is a finite, but very small probability of finding two structures with the same InChIKey. However the probability for duplication of only the first block of 14 characters has been estimated as one duplication in 75 databases each containing one billion unique structures; such duplication therefore appears unlikely at present.
The nomenclature of ChEBI was extended with the capture of the INN for pharmaceutical substances. The INN is the official non-proprietary or generic name given to a pharmaceutical substance, as designated by the World Health Organization (WHO). INNs may appear in ChEBI in English, Latin, Spanish and French language versions. ChEBI now captures synonyms in English, Latin, Spanish, German and French.
Availability
All data in the database and on the FTP server are non-proprietary or are derived from a non-proprietary source. It is thus freely accessible and available to anyone. In addition, each data item is fully traceable and explicitly referenced to the original source. Apart from web access, the entire ChEBI data are provided in four different formats and can be downloaded from the FTP server (ftp://ftp.ebi.ac.uk/pub/databases/chebi/).
Flat-file table dumps
ChEBI is stored in a relational database and we currently provide the ChEBI tables in a flat-file tab delimited format. The files are stored in the same structure as the relational database.
Oracle binary table dumps
ChEBI provides an Oracle binary table dump that can be imported into an Oracle relational database.
Generic structured query language table dumps
ChEBI provides a generic structured query language (SQL) dump which consists of SQL insert statements. These insert statements should be usable in any database which accepts SQL as its query language.
OBO ontology format
ChEBI provides the ChEBI ontology in OBO format version 1.2 (http://www.geneontology.org/GO.format.obo-1_2.shtml).
SD file format
ChEBI provides the chemical structures in MDL SD file format as well as associated data.
All download files are available in two flavours, namely downloads with only three star entities or downloads with both three and two star entries.
CONCLUSION
Since our first report on the database and ontology of ChEBI in 2007, the resource has grown to >400 000 molecules, owing to the incorporation of compound data from the manually annotated chemogenomics database ChEMBL which has been established at the EBI. We have made substantial improvements to the data quality of ChEBI via additional data annotations as well as extentions of the ChEBI ontology. Submitters have contributed 202 new entities (July 2009) since the introduction of the submission tool in May 2009. We expect this to increase with the introduction of a new feature allowing update of existing ChEBI entities.
Additional cross-references to patent data as well as to resources such as BRENDA and NMRShiftDB have allowed users to use ChEBI as a chemistry portal to other resources. ChEBI now has >5 000 000 cross-references to other resources. We expect this to grow as additional data are updated from submitters and the ChEMBL database.
FUNDING
European Commission [FELICS 021902, SLING 226073] and the Biotechnology and Biological Sciences Research Council [BB/G022747/1]. Funding for open access charge: European Commission [SLING 226073].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the contribution of the following people to the success of the project: Michael Ashburner, Kristian Axelsen, Colin Batchelor, Peter Corbett, Hélène Courrier, Kirill Degtyarenko, Stefan Kuhn, Anne Morgat, Jeremy Parsons, Mark Rijnbeek, John Overington and the ChEMBL team as well as all ChEBI submitters. They are also grateful for the software contribution of ChemAxon.
REFERENCES
- 1.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimitropoulos D, Ionides J, Henrick K. Using MSDchem to Search the PDB Ligand Dictionary. Curr. Protoc. Bioinform. 2006:14.3.1–14.3.21. doi: 10.1002/0471250953.bi1403s15. [DOI] [PubMed] [Google Scholar]
- 3.Batchelor C. An upper-level ontology for chemistry. In: Eschenbach C, Grüninger M, editors. Formal Ontology in Information Systems, Proceedings of the Fifth international Conference, FOIS 2008, Saarbrücken, October 31st – 3rd November 2008. Amsterdam: IOS Press; 2008. pp. 195–207. [Google Scholar]
- 4.Smith B, Cuesters W, Klagges B, Köhler J, Kumar A, Lomax J, Mungall C, Neuhaus F, Rector AL, Rosse C. Relations in Biomedical Ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinbeck C, Kuhn S. NMRShiftDB—compound identification and structure elucidation support through a free community-built web database. Phytochemistry. 2004;65:2711–2717. doi: 10.1016/j.phytochem.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rijnbeek M, Steinbeck S. An open source chemistry search engine for Oracle. J. Cheminf. doi: 10.1186/1758-2946-1-17. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinbeck C, Hoppe C, Kuhn S, Floris M, Guha R, Willighagen EL. Recent Developments of the Chemistry Development Kit (CDK)—an open-source Java library for chemo- and bioinformatics. Curr. Pharm. Des. 2006;12:2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- 9.Chang A, Scheer M, Grote A, Schomburg A, Schomburg D. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:D588–D592. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcántara R, Ast V, Axelson KB, Darsow M, de Matos P, Ennis M, Morgat A, Degtyarenko K. IntEnz. Molecular Biology Database Collection entry number 508. Nucleic Acids Res. 2007 http://www.oxfordjournals.org/nar/database/summary/508. [Google Scholar]
- 11.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update—from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2008;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Novère N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, Li L, Sauro H, Schilstra M, Shapiro B, et al. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34:D689–D691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittig U, Golebiewski M, Kania R, Krebs O, Mir S, Weidemann A, Anstein S, Saric J, Rojas I. Proceedings of the 3rd International workshop on data integration in the life sciences (DILS'0;6), Hinxton, UK. Lecture Notes in Bioinformatics, 4075. Berlin and Heidelberg: Springer; 2006. SABIO-RK: Integration and curation of reaction kinetics data; pp. 94–103. [Google Scholar]
- 15.Corbett P, Murray-Rust P. High-throughput identification of chemistry in life science texts. In: Berthold MR, Glen RC, Fischer I, editors. Computational Life Sciences II, Second International Symposium, CompLife 2006, Cambridge, UK, September 27-29, 2006, Proceedings. Lecture Notes in Computer Science 4216. Berlin and Heidelberg: Springer; 2006. pp. 107–118. [Google Scholar]
- 16.Stein SE, Heller SR, Tchekhovski D. Proceedings of the 2003 International Chemical Information Conference (Nimes) Tetbury, UK: Infonortics; 2003. An open standard for chemical structure representation: the IUPAC Chemical Identifier; pp. 131–143. [Google Scholar]