Abstract
The University of California, Santa Cruz (UCSC) Genome Browser website (http://genome.ucsc.edu/) provides a large database of publicly available sequence and annotation data along with an integrated tool set for examining and comparing the genomes of organisms, aligning sequence to genomes, and displaying and sharing users’ own annotation data. As of September 2009, genomic sequence and a basic set of annotation ‘tracks’ are provided for 47 organisms, including 14 mammals, 10 non-mammal vertebrates, 3 invertebrate deuterostomes, 13 insects, 6 worms and a yeast. New data highlights this year include an updated human genome browser, a 44-species multiple sequence alignment track, improved variation and phenotype tracks and 16 new genome-wide ENCODE tracks. New features include drag-and-zoom navigation, a Wiki track for user-added annotations, new custom track formats for large datasets (bigBed and bigWig), a new multiple alignment output tool, links to variation and protein structure tools, in silico PCR utility enhancements, and improved track configuration tools.
INTRODUCTION
The University of California, Santa Cruz (UCSC) Genome Browser (1,2) is a web-based resource for the biomedical community, providing timely and convenient access to sequence and annotations for human and other vertebrate reference species genomes, along with selected model invertebrates. The minimal set of annotation ‘tracks’ on every browser generally includes assembly and gaps, the percentage of guanine and cytosine bases, alignments of RefSeq genes (3,4), mRNAs and ESTs obtained from GenBank (5) and repeat region annotations. Most browsers also include one or more gene or gene prediction tracks, including Ensembl Genes (6), as well as comparative genomics tracks that show pairwise genomic sequence alignments (chain and net tracks) between organisms. About half of the organisms hosted in the browser include multiple sequence alignment (multiz) tracks (7). The human and mouse browsers feature a gene prediction track created at UCSC—UCSC Genes (2,8,9)—based on data from RefSeq, GenBank, CCDS and UniProt. Many more expression, regulation, variation and phenotype tracks are available on the model organism and human browsers.
Track documentation is accessible by clicking on a track feature, the track name, or the vertical bar directly to the left of the track in the graphical display. We also provide links from the browser to the corresponding locations on the NCBI Map Viewer (10) and Ensembl (6) genome browsers.
UCSC is the Data Coordination Center for the Encyclopedia of DNA Elements (ENCODE) project (11,12). This project, which aims to identify all functional elements in the human genome sequence, uses the Genome Browser as the primary data portal. ENCODE pilot phase data covering ∼1% of the human genome is available on the hg17 and hg18 (NCBI build 35 and 36) human genome browsers; genome-wide, production phase data is available on the hg18 assembly.
The Genome Browser website offers several tools for visualizing and analyzing genome sequences and annotation tracks. Both BLAT (13) and the in silico PCR tool quickly map sequences to genomes. The LiftOver utility translates genomic coordinates between assemblies and is available in a web interface version (at http://genome.ucsc.edu/cgi-bin/hgLiftOver) and as a command-line executable program (from http://hgdownload.cse.ucsc.edu/admin/exe/). The Table Browser (14) provides a graphical interface for retrieving, filtering, intersecting, correlating and summarizing data from Genome Browser database tables. The Gene Sorter (15) allows for exploration of gene relationships by several metrics, such as expression profiles and protein homology, and displays many types of data for each gene. Protein sequence properties are displayed as tracks and histograms in the Proteome Browser (16). VisiGene (17) displays a large collection of Xenopus and mouse in situ images that show gene expression patterns. The custom tracks tool allows the upload of user data as tracks in the browser for visualization in the context of other annotation tracks and for manipulation with the Table Browser. Custom tracks and configuration settings can be saved and shared with the Session (9) tool. Genome Graphs (9) provides a genome-wide view of both hosted tracks and user-generated custom tracks. The Genome Browser tools are integrated with one another, with links provided between tools.
Instructions for using all of the Genome Browser tools are provided via a context-sensitive ‘Help’ link at the top of every page and a ‘Training’ link on the homepage (http://genome.ucsc.edu/training.html). Free tutorials produced by OpenHelix are available from http://www.openhelix.com/ucsc/. Our FAQ (http://genome.ucsc.edu/FAQ/) provides answers to frequently asked questions, and new questions are answered through the searchable archives of our public mailing lists (http://genome.ucsc.edu/contacts.html) and directly from our staff (see ‘Contacting Us’ section). Other useful information contributed by the browser staff and users is available on the UCSC Genome Browser wiki site at http://genomewiki.ucsc.edu/.
The server at http://hgdownload.cse.ucsc.edu/ provides bulk downloads of the browser sequence and annotation data, as well as the Genome Browser source code. The source can be used to set up a mirror site (see instructions at http://genome.ucsc.edu/admin/mirror.html) or to install the large collection of browser bioinformatics command-line utilities on a local computer (see a list at http://genomewiki.ucsc.edu/index.php/Kent_source_utilities). Mirrors of the Genome Browser need not host all of the data hosted by UCSC; assemblies and data of interest can be selectively mirrored (see http://genomewiki.ucsc.edu/index.php/Minimal_Browser_Installation). Users may be interested in setting up a mirror site to host confidential data, to display customized browsers and tracks, and for faster data access.
NEW DATA
New assemblies
Since September 2008, we have updated the genome assemblies for horse, human, opossum, medaka and yeast. The new human assembly, UCSC version hg19 (Genome Reference Consortium GRCh37), includes pairwise alignments to 4 primates, 7 non-primate placental mammals and 12 non-placental vertebrates, and we plan to add a 46-species Conservation track by early 2010. We expect the set of hg19 annotations to grow substantially during the next year as new tracks are added and ENCODE datasets are migrated from the hg18 human assembly.
Regular updates to existing tracks
Several browser tracks are updated on a regular basis. The RefSeq and mRNA tracks, which show sequences from all organisms in GenBank aligned to all of the Genome Browser assemblies, are updated nightly. The RefSeq tracks now include alignments of non-coding genes. EST tracks are updated with new data deposited in GenBank on a weekly basis. Other tracks that are updated each night include the Mammalian Gene Collection (MGC) tracks (18) on the human, mouse, rat, cow and frog browsers, the Consensus Coding Sequence (CCDS) tracks (19) on the human and mouse, and the ORFeome Clones tracks (which show alignments of clones from the ORFeome Collaboration) on the human genome. The Ensembl Genes (6) track, available on ∼25 different organisms, and the Database of Genomic Variants (DGV) track (20,21), which contains genomic variations observed in healthy human individuals, are updated whenever a new version is released.
On mouse browsers, tracks that show alignments of sequence tags from the International Gene Trap Consortium (IGTC) (22) are updated monthly. Mouse Genome Informatics (MGI) (23) tracks, which show quantitative trait loci, phenotypes and alleles; and the IKMC Genes track, which shows genes targeted by the International Knockout Mouse Consortium (24) for generating mouse embryonic stem cells containing a null mutation in every gene in the mouse genome, are all updated regularly.
New annotation tracks
In the past year, ∼100 new annotation tracks have been added to the Genome Browser in more than 1000 tables, of which some 500 were released as part of 16 new ENCODE tracks on the hg18 human browser. (See Rosenbloom et al., this issue, for detailed information on ENCODE tracks). The remainder of this section describes some of these new additions to the browser.
44-species vertebrate alignment and conservation
In January 2009, we released a new Conservation (25) track for the human hg18 assembly that displays multiz (7) multiple sequence alignments of 43 vertebrate species to the human genome, plus two different measurements of evolutionary conservation. In addition to the assemblies included in the previous (28-species) Conservation track (9), we have added four new high-coverage (>6×) assemblies: orangutan, marmoset, zebra finch and lamprey; and 12 new lower-coverage assemblies: gorilla, tarsier, mouse lemur, kangaroo rat, squirrel, pika, alpaca, dolphin, microbat, megabat, rock hyrax and sloth.
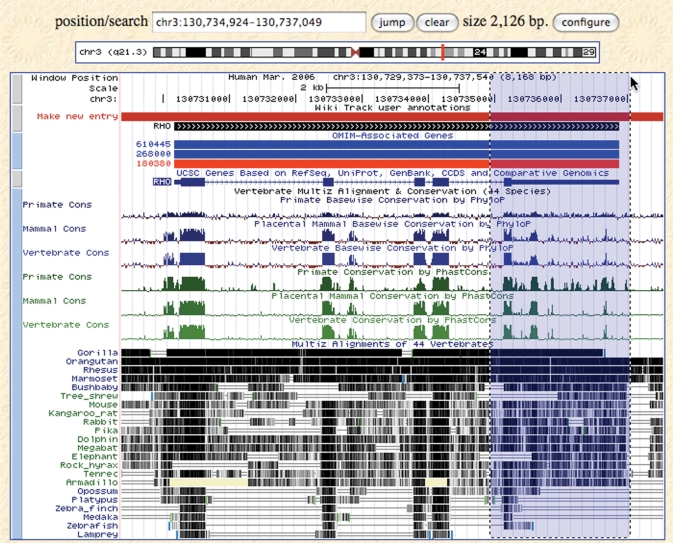
Both phastCons (25) and phyloP (26) conservation scores have been computed separately for three groups of organisms: 8 primates alone, 31 placental mammals, and all 44 vertebrates. The two conservation scores are informative in different ways. The phastCons score takes neighboring bases into account, estimating the probability that each nucleotide belongs to a conserved element. It is sensitive to ‘runs’ of conserved sites and is used to create the Conserved Elements subtrack of the Conservation track. The phyloP score is a separate measurement of conservation at each base, ignoring neighboring bases in its calculation. It can measure acceleration (faster evolution than expected under neutral drift) as well as conservation (slower than expected evolution). PhyloP is useful for evaluating signatures of selection at particular nucleotides (e.g. third codon positions, or first positions of miRNA target sites). Figure 1 shows a subset of species in the 44-way multiple alignment track and all six conservation score tracks.
Figure 1.
Main Genome Browser display page on the hg18 human assembly, showing the Wiki track, OMIM Genes, UCSC Genes and the 44-species Conservation track, with all six conservation score tracks visible. The highlighted region shows drag-and-zoom in action. When the mouse button is released, the browser will redraw at the new coordinates listed in the position/search box.
OMIM genes
A new OMIM Genes track in the Phenotype and Disease Associations section of the human browser displays the positions of genes annotated in the Online Mendelian Inheritance in Man (OMIM) database (27). This augments the existing OMIM linkouts on the details pages of the UCSC Genes track with a quick visual way to identify the locations of these annotations. The OMIM Genes track shows the locations of UCSC Genes that have associated OMIM identifiers mapped by RefSeq (4) and UniProt (28,29). If the gene is associated with a disease phenotype in OMIM, the browser displays it in red; otherwise, it is displayed in blue (Figure 1).
Variation
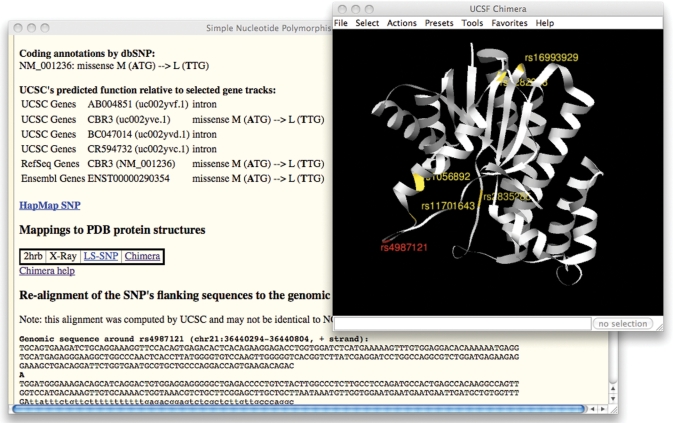
We have added a SNP track to the hg18 human browser for dbSNP (30) build 130. The track contains close to 19 million SNP annotations, and includes the first set of SNP calls from the 1000 Genomes Project (31). About 5.6 million reference SNPs in build 130 include some evidence submitted by 1000 Genomes; of those SNPs, nearly 1 million identifiers are newly added since dbSNP build 129. We have also added the option to display each SNP’s predicted functional role (such as coding-non-synonymous, coding-synonymous, intron, splice site, etc.) relative to different gene sets on the SNP details pages (Figure 2).
Figure 2.
Details page of SNP rs4987121 from the SNPs (130) track, showing the new ‘predicted function relative to selected gene tracks’ feature using UCSC Genes, RefSeq Genes and Ensembl Genes. The window on the right is the Chimera 3D protein structure viewer, opened by clicking on the ‘Chimera’ link in the first window. The selected SNP is highlighted in red, while other non-synonymous SNPs are highlighted in gold.
There are several new tracks of data from the Human Genome Diversity Project (HGDP) (32), which assayed 660 000 SNPs in individuals from 53 populations worldwide. The browser shows per-population allele frequencies plotted on a world map on the details page for each HGDP SNP. In addition, HGDP provided several per-continent population genetics measures: FST, heterozygosity, iHS and XP-EHH.
A new ‘Genome Variants’ track on the hg18 human browser shows single-nucleotide differences from several published personal genome sequences. We plan to add variants found in the 1000 Genomes Project’s six high-coverage individuals to this track in the near future, as well as additional published genomes.
Expression
We have added a number of new probesets to our collection of gene chip mappings. These datasets show different commercially available probesets mapped together on the genome assembly. These are useful to researchers designing experiments, as the coverage and locations of probes from the many sources are collected in one convenient location in the graphical display. In the hg18 human browser, the probesets are configured together in a composite track. The mappings are easily downloaded in whole or in part via the Table Browser.
NEW FEATURES
Drag-and-zoom
The drag-and-zoom feature enables users to quickly zoom in to areas of interest in the Genome Browser. The feature is accessed by clicking on the desired start coordinate in the Base Position track and then dragging the mouse to the desired end coordinate to define a new position. The coordinate range in the position/search box is dynamically updated as the mouse is moved, and the browser image redraws at the new coordinates when the mouse button is released (Figure 1).
Annotating the genome using the Wiki track
Users are now encouraged to contribute information to the Genome Browser via the Wiki track, available in the Mapping and Sequencing Tracks section on the hg18 and hg19 human browsers and the mm9 mouse browser. The Wiki track contains user-added annotations that are displayed like regular annotations; clicking on an item in the track opens a corresponding page for the gene or region on the Genome Browser wiki site (http://genomewiki.cse.ucsc.edu/), where any user can log in and create or edit pages (9). Wiki track annotations may be added for a specific gene locus in the UCSC Genes track by clicking on a gene and then the ‘User annotations’ link. Alternatively, annotations to any genomic region within a chromosome may be made by clicking on the red ‘Make new entry’ bar at the top of the Wiki track. This interactive mode of adding content to the browser allows researchers to directly annotate genes in their areas of expertise.
Custom tracks for large datasets
To improve display performance of custom tracks based on very large datasets, we have added functionality to let users store data files locally on their own web-accessible server (http, https, or ftp) and then view them as custom tracks in the Genome Browser without uploading the entire files to UCSC. The data files are stored in one of two new formats: bigBed or bigWig. These are binary formats that use an R-tree internally to index genome coordinate ranges, making it possible to selectively transfer only the subset of data needed to display regions actually viewed in the Genome Browser.
To create a file in one of the new ‘big’ formats, users start with a BED, BedGraph, or Wiggle text file and then run a conversion program on their own machine to convert the text file to the corresponding binary format. Executable versions of the conversion programs, called bedToBigBed, bedGraphToBigWig and wigToBigWig, are available on the Genome Browser downloads server (http://hgdownload.cse.ucsc.edu/admin/exe/), along with several programs to aid in working with the new binary data file types. Once a bigBed or bigWig file is viewed as a custom track, the uploaded data remains cached at UCSC for at least 48 h from last use without any additional data transfer. The life of the cached data can be extended by creating a session (9) that includes the ‘big’ custom track.
Bridging genomic variation and protein structure
To assist in exploring the impact of variation on protein function, we have added functionality to connect genes and observed genomic variation to protein structures. Two new related tools for visualizing non-synonymous SNPs mapped to proteins with solved structures are accessible from the UCSC Genes and SNP track details pages via ‘LS-SNP’ and ‘Chimera’ links.
Clicking an LS-SNP link will open a corresponding SNP or protein page in LS-SNP/PDB (33), a web tool that provides mappings of human non-synonymous SNPs onto protein structures deposited in the Protein Data Bank (34). It provides information useful for identifying the non-synonymous SNPs that are most likely to have an impact on biological function.
UCSF Chimera (35) is a 3D protein structure viewer that runs as a web browser helper application on a user’s computer. Once the Chimera application is downloaded and installed, clicking a Chimera link in the Genome Browser will launch the program and display the selected 3D protein structure with all of the non-synonymous SNP residues (identified by LS-SNP/PDB) colored gold and labeled with the dbSNP identifier. When launching Chimera from a SNP track details page, the residue of the selected SNP will be colored red (Figure 2).
In silico PCR for transcribed sequences
The in silico PCR tool lets users submit a pair of PCR primers and returns the target sequence that those primers will produce. The initial version of this tool worked only on genomic sequence, but has since been extended on the human and mouse browsers to allow the query of transcribed sequences as well. When ‘UCSC Genes’ is selected as the target, primers that occur within the exonic regions of a gene, even those spanning long introns, will produce PCR product sequence for all gene isoforms with the introns removed. Clicking the links on the results page displays graphical representations of the PCR products in the browser, with the location of the primers indicated.
CDS FASTA alignments
The Conservation track in the Comparative Genomics section of the browser shows genomic sequence alignments of multiple species. We have provided a way to display these multiple alignments for coding regions of genes, either as nucleotides or as amino acids in FASTA format. The tool is available via the Table Browser and from the details pages of the UCSC Genes and RefSeq Genes tracks. In the Table Browser, when a gene prediction track is selected and a multiple alignment is available for the selected species, the option for ‘CDS FASTA alignment from multiple alignment’ appears in the output format menu. Several options are available for formatting the output, including the option to exclude species from the multiple alignment display.
Improved track configuration
The multi-view display functionality developed for the ENCODE project (Rosenbloom, et al., this issue) is now used in several other tracks, including the new 44-species conservation track on the hg18 human browser and the chain and net tracks on the hg19 human browser. This functionality makes it possible to manipulate the display of related subtracks within a large track at several levels: by data type, by specific track attributes, or at the individual track level. Additionally, the track display order on the main browser page can be controlled for multi-view tracks. For example, the subtracks within the Primate Chain/Net track on hg19 can be configured to group together all tracks for a given primate, all of the chain tracks, or all tracks for a particular clade (Hominidae, Cercopithecinae or Haplorrhini). This configurability gives users greater flexibility to create customized displays.
Performance enhancements
In September 2009, we made several changes to underlying Genome Browser code to speed up the track display, especially when large chromosomal regions are viewed. We also upgraded and expanded the hardware that serves the MySQL tables in the Genome Browser database, which has improved interactive response times, and have upgraded from MySQL version 4.0 to version 5.0.
FUTURE DIRECTIONS
We will continue to incorporate new and updated vertebrate and selected model invertebrate assemblies to the UCSC Genome Browser. We expect elephant, rabbit, sea hare and updated zebrafish and Tetraodon browsers to be available on our site by early 2010. We anticipate several types of variation data from the 1000 Genomes Project, including SNPs and copy number variants (CNVs) called from pilot project data. [The first round of SNPs submitted by 1000 Genomes in December 2008 is already available in the ‘SNPs (130)’ track.] Further improvements to the UCSC Genes track are planned.
New features will include support for Binary Sequence Alignment/Map (BAM) (36) as a custom track file format so that high-coverage sequencing read alignments from 1000 Genomes and other sequencing projects can be displayed in the browser. This will make it possible to view the underlying data from which SNPs and CNVs were called. We plan to investigate high performance solutions for mapping, visualizing and analyzing next-generation sequencing data. We are working on enhancements to the Genome Browser user interface: drag-and-drop track reordering, functionality to navigate the main browser display by horizontal drag-scrolling, and a new track-search interface to help users locate data useful to them.
CONTACTING US
Questions and feedback about the Genome Browser may be directed to our public, moderated mailing list at genome@soe.ucsc.edu. A separate mailing list is maintained for questions pertaining to mirroring the browser at genome-mirror@soe.ucsc.edu. Note that all correspondence sent to either of the afore-mentioned addresses is available on a public, searchable archive, and individual messages cannot be removed. Users are encouraged to browse and search the archives of previously answered questions from http://genome.ucsc.edu/contacts.html before submitting new questions. Reports of problems accessing the browser and questions not appropriate for the public forum can be directed to genome-www@soe.ucsc.edu. We announce items such as new genome assembly releases, new software features, and planned server downtime on genome-announce@soe.ucsc.edu; visit https://lists.soe.ucsc.edu/mailman/listinfo/genome-announce to subscribe.
FUNDING
National Human Genome Research Institute (5P41HG002371-09 and 5U41HG004568-02 to D.H., subcontract on 1U54HG004555-01 to T.H. at the Wellcome Trust Sanger Institute); the Howard Hughes Medical Institute (to D.H.); the National Institute of Diabetes and Kidney Diseases, National Institutes of Health (2 R01 DK065806-06 to R.H. at The Pennsylvania State University). Funding for open access charge: The Howard Hughes Medical Institute.
Conflict of interest statement. B.R., D.K., R.M.K., A.S.H., A.S.Z., P.A.F., M.D., K.E.S., K.R.R., B.J.R., A.P., M.P., L.R.M., F.H., R.A.H., T.R.D., H.C., G.P.B., D.H. and W.J.K. receive royalties from the sale of UCSC Genome Browser source code licenses to commercial entities.
ACKNOWLEDGEMENTS
We wish to thank the numerous collaborators worldwide who have contributed annotation data to the Genome Browser, the individuals on our Scientific Advisory Board who guide our work, and our users, who continually give valuable feedback and support. We would also like to acknowledge Jorge Garcia, Erich Weiler, Victoria Lin and Alex Wolfe, our dedicated team of system administrators, for providing an outstanding computing environment.
REFERENCES
- 1.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith K, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 9.Karolchik D, Kuhn R, Baertsch R, Barber G, Clawson H, Diekhans M, Giardine B, Harte R, Hinrichs A, Hsu F, et al. The UCSC genome browser database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas DJ, Rosenbloom KR, Clawson H, Hinrichs AS, Trumbower H, Raney BJ, Karolchik D, Barber GP, Harte RA, Hillman-Jackson J, et al. The ENCODE Project at UC Santa Cruz. Nucleic Acids Res. 2007;35:D663–D667. doi: 10.1093/nar/gkl1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 13.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent WJ, Hsu F, Karolchik D, Kuhn RM, Clawson H, Trumbower H, Haussler D. Exploring relationships and mining data with the UCSC Gene Sorter. Genome Res. 2005;15:737–741. doi: 10.1101/gr.3694705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu F, Pringle TH, Kuhn RM, Karolchik D, Diekhans M, Haussler D, Kent WJ. The UCSC Proteome Browser. Nucleic Acids Res. 2005;33:D454–D458. doi: 10.1093/nar/gki100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, et al. The UCSC Genome Browser Database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P, et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;18:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 21.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 22.Nord AS, Chang P, Conklin B, Cox A, Harper C, Hicks GG, Huang CC, Johns SJ, Kawamoto M, Liu S, et al. The International Gene Trap Consortium Website: a portal to all publicly available gene trap cell lines in mouse. Nucleic Acids Res. 2006;34:D642–D648. doi: 10.1093/nar/gkj097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA Mouse Genome Database Group. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Comprehensive Knockout Mouse Project Consortium. Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, et al. The Knockout Mouse Project. Nat. Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou MM, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siepel A, Pollard KS, Haussler D. Proceedings of the 10th International Conference on Research in Computational Molecular Biology (RECOMB 2006: April 2–5, 2006, Venice Lido, Italy) 2006. New methods for detecting lineage-specific selection; pp. 190–205. [Google Scholar]
- 27.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's; Online Mendelian Inheritance in Man (OMIM®) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The UniProt Consortium. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherry S, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski E, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaiser J. A plan to capture human diversity in 1000 genomes. Science. 2008;319:395. doi: 10.1126/science.319.5862.395. [DOI] [PubMed] [Google Scholar]
- 32.Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan M, Diekhans M, Lien S, Liu Y, Karchin R. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics. 2009;25:1431–1432. doi: 10.1093/bioinformatics/btp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berman H, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen E, Goddard T, Huang C, Couch G, Greenblatt D, Meng E, Ferrin T. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]