Abstract
SoyBase, the USDA-ARS soybean genetic database, is a comprehensive repository for professionally curated genetics, genomics and related data resources for soybean. SoyBase contains the most current genetic, physical and genomic sequence maps integrated with qualitative and quantitative traits. The quantitative trait loci (QTL) represent more than 18 years of QTL mapping of more than 90 unique traits. SoyBase also contains the well-annotated ‘Williams 82’ genomic sequence and associated data mining tools. The genetic and sequence views of the soybean chromosomes and the extensive data on traits and phenotypes are extensively interlinked. This allows entry to the database using almost any kind of available information, such as genetic map symbols, soybean gene names or phenotypic traits. SoyBase is the repository for controlled vocabularies for soybean growth, development and trait terms, which are also linked to the more general plant ontologies. SoyBase can be accessed at http://soybase.org.
INTRODUCTION
The last decade has seen a significant increase in soybean [Glycine max (L.) Merr.] research. The first molecular genetic map of only a few hundred RFLP markers has grown to over 4000 loci encompassing RFLP, RAPD, SSR and SNP markers (1). Over a thousand quantitative trait loci (QTL) representing more than 90 agronomically important traits have been mapped in soybean. More than 1.4 million nucleotide and expressed sequence tag (EST) sequences are available in public repositories. Macro- and micro-arrays based on ESTs have been developed and are being used to generate expression data for thousands of genes under different experimental conditions. The recently completed initial assembly of the genomic sequence of the cultivar ‘Williams 82’ is available (Schmutz et al., in preparation). Sequence annotation tracks for gene calls, the BAC-based physical map (2), the Affymetrix SoyChip1 probe sets and numerous gene expression projects are provided in SoyBase. Based on the needs of the soybean research community, the USDA-ARS developed SoyBase as a central repository for genetic and genomic data and related resources for soybean, as well as a single starting point for access to other laboratory-specific web pages and specialized data sets. In this article we present an overview of the major sections of SoyBase and some of the tools available for data mining and searching the database.
RESULTS AND DISCUSSION
General directions for using SoyBase
SoyBase is organized into four broad sections (Figure 1):
genetic, QTL and physical maps
genome sequence and annotations
analysis tools and data mining tools
data and community resources
Figure 1.
The main navigation aids that are present on all SoyBase pages, and which allow the user to quickly move between the sections of the web site. The main tabs (i.e. Maps or Resources) link immediately to the selected page. Entries on the second line can be used to move directly to a specific SoyBase page.
Genetic and physical maps
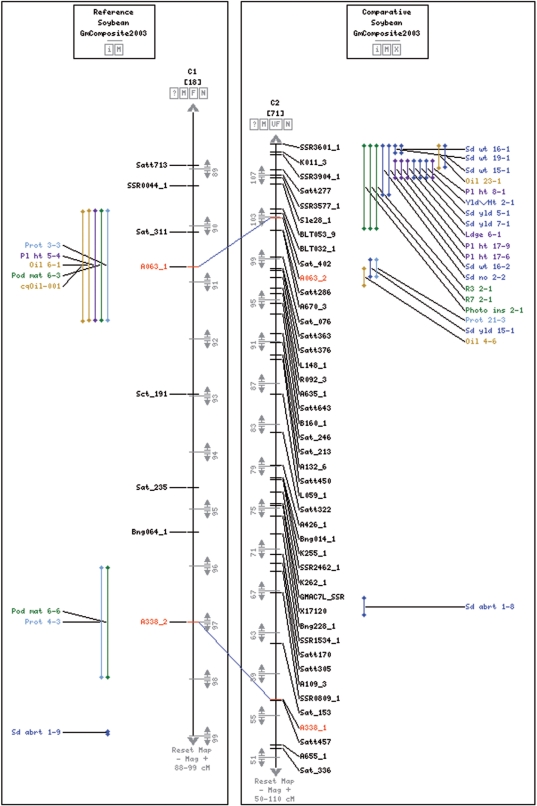
The genetic and physical (FPC) maps in SoyBase are displayed using the comparative map viewer CMap (a component of Generic Model Organism Database Project; http://gmod.org; (3)). In addition to providing views of single linkage groups, CMap provides the ability to simultaneously view related maps. Soybean is a recent tetraploid, having undergone polyploidy an estimated 10–15-million years ago (4,5). The ability to view multiple evolutionarily related chromosomes or regions is, therefore, particularly useful in soybean. Figure 2 shows an example of genetic maps from related regions of two homoeologous chromosomes.
Figure 2.
Homoeologous regions of soybean linkage groups C1 and C2 (chromosomes Gm04 and Gm06). Broad QTL classes are indicated by color. Only a subset of the QTL are shown so that potentially related QTL can easily be recognized.
All map features are linked to extensive textual data in SoyBase. Genetic markers have been positioned on both the genetic and sequence maps, thus providing facile movement between these two views of the soybean genome. Contextual menus are used to access the textual data and to move between the genetic and genome sequence views of the chromosomes.
Genomic sequence
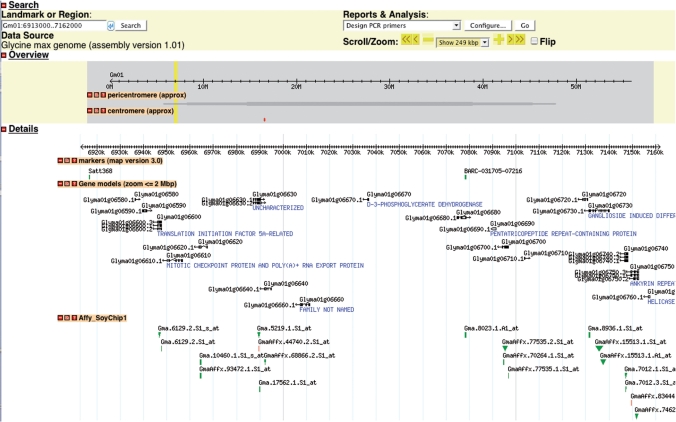
The ‘Williams 82’ genomic sequence and associated features are accessible using the GBrowse genome viewer (http://gmod.org). The genomic sequence, sequenced by the DOE-JGI and assembled in 2008 by a multi-agency consortium, spans nearly 1 billion bases (Soybean Genome Sequencing Consortium, http://www.phytozome.net/soybean.php; and Schmutz et al., in preparation). Figure 3 shows a region of chromosome Gm01 with some of the annotation tracks that are available. Contextual menus provide additional information for sequence features and links between the sequence and genetic maps.
Figure 3.
Genome sequence view of a region of soybean chromosome Gm01. The tracks shown here are genetic markers, gene models from the 1.01 annotation (Soybean Genome Sequencing Consortium), and locations of Affymetrix probesets. For each of these tracks, a click on a feature provides a contextual menu with relevant links: to CMap from markers; to several sequence tools from gene models; and to PLEXdb (6) from Affymetrix probe sets. Many other annotation tracks are available, including intragenomic synteny blocks, the BAC-based FPC contigs that comprise the physical map, all of the fingerprinted and end-sequenced BACs, and EST contigs from both soybean and other legumes.
The number of studies on gene expression using either the Affymetrix SoyChip1 or next generation sequencing strategies is rapidly increasing. To accommodate these data we have partnered with PLEXdb (http://plexdb.org; Wise et al., 2007). In addition a subset of the short read (i.e. 454 or Solexa) expression data, as well as the Affymetrix probe sets, are presented in the SoyBase genome sequence viewer. Links are provided to PLEXdb to allow the user to further explore the experimental design and data. Links are also available from PLEXdb to SoyBase to allow the experimental results to be analyzed in the context of the genetic and genomic data in SoyBase.
Tools
SoyBase contains a number of analysis tools, including
-
– Sequence similarity searches of Glycine max and G. soja sequences:
BLAST and BLAT can be used to search all or a subset of the Glycine sequences (ESTs, gene indices, etc.), including the genome sequence. The results of a BLAT search against the whole genome sequence are shown directly in the genome sequence browser.
-
–Search Glycine max and G. soja EST sequence libraries by keywords:
Text based searches of the library and individual EST annotations can be performed. Results are returned in a format suitable for viewing in a web browser or for import into other analysis programs or databases.
-
–Search the annotations for Affymetrix SoyChip contig sequences:
A comprehensive annotation has been developed for each of the probe sets on the Affymetrix SoyChip. A user can provide a file of probe set names of interest and get a report containing their annotations.
-
–Search for unigene sequences that match Affymetrix SoyChip probe sequences:
Several unigene sets have been constructed for soybean ESTs using different assembly parameters. This tool allows a user to get the unigene(s) associated with one or more probe sets from each of the unigene assemblies.
-
–Browse or search soybean ontologies:
SoyBase contains the most complete trait, growth and developmental ontologies available. These ontologies were developed by SoyBase staff to suggest a controlled vocabulary for soybean field growth stages (SoyWGR), individual plant development (SoyGRO) and traits (SoyTO). Where applicable, soybean specific terms have been associated with their Plant Ontology (PO) and Gramene Plant Trait Ontology (TO) synonyms to facilitate cross species comparisons.
Resources
SoyBase is intended to be a central resource for soybean researchers. In addition to the data, maps and tools described above, a number of community resources are made available either as data or as links to other sites. These include, among others, links to other soybean-centric websites and laboratories, a list of upcoming meetings of potential interest to soybean researchers and links to an extensive collection of USDA sites about soybean breeding and production.
Data availability
All of the data in SoyBase is freely available. Please use the ‘Contact Us’ page at SoyBase or email the curator (David Grant, david.grant@ars.usda.gov) to request any specific subset of the data.
FUTURE PLANS
SoyBase is continuously updated to include new data as they become available. In addition new data types are incorporated and linked to the existing data when appropriate. Some new data types that will soon be added to SoyBase include
– allele data and frequencies for the genetic markers.
–genetic marker-based haplotypes for the soybean germplasm collection.
–integrated maps and data for the identified retrotransposon and DNA transposable elements in soybean.
FUNDING
United States Department of Agriculture, Agricultural Research Service (USDA-ARS). Funding for open access charge: USDA-ARS.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The SoyBase Development Group includes David Grant, Rex Nelson, Steven Cannon, Nathan Weeks, Andrew Denner and Randy Shoemaker.
REFERENCES
- 1.Choi IY, Hyten DL, Matukumalli LK, Song Q, Chaky JM, Quigley CV, Chase K, Lark KG, Reiter RS, Yoon MS, et al. A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics. 2007;176:685–696. doi: 10.1534/genetics.107.070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shoemaker RC, Grant D, Olson T, Warren WC, Wing R, Yu Y, Kim H, Cregan P, Joseph B, Futrell-Griggs M, et al. Microsatellite discovery from BAC end sequences and genetic mapping to anchor the soybean physical and genetic maps. Genome. 2008;51:294–302. doi: 10.1139/G08-010. [DOI] [PubMed] [Google Scholar]
- 3.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC. Mining EST databases to resolve evolutionary events in major crop species. Genome. 2004;47:868–876. doi: 10.1139/g04-047. [DOI] [PubMed] [Google Scholar]
- 5.Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ. Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst. Biol. 2005;54:441–454. doi: 10.1080/10635150590945359. [DOI] [PubMed] [Google Scholar]
- 6.Wise RP, Caldo RA, Hong L, Shen L, Cannon EK, Dickerson JA. BarleyBase/PLEXdb: a unified expression profiling database for plants and plant pathogens. In: Edwards D, editor. Methods in Molecular Biology. Plant Bioinformatics—Methods and Protocols. Vol. 406. Totowa, NJ: Humana Press; 2007. pp. 347–363. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the data in SoyBase is freely available. Please use the ‘Contact Us’ page at SoyBase or email the curator (David Grant, david.grant@ars.usda.gov) to request any specific subset of the data.