Abstract
The laboratory mouse is the premier animal model for studying human disease and thousands of mutants have been identified or produced, most recently through gene-specific mutagenesis approaches. High throughput strategies by the International Knockout Mouse Consortium (IKMC) are producing mutants for all protein coding genes. Generating a knock-out line involves huge monetary and time costs so capture of both the data describing each mutant alongside archiving of the line for distribution to future researchers is critical. The European Mouse Mutant Archive (EMMA) is a leading international network infrastructure for archiving and worldwide provision of mouse mutant strains. It operates in collaboration with the other members of the Federation of International Mouse Resources (FIMRe), EMMA being the European component. Additionally EMMA is one of four repositories involved in the IKMC, and therefore the current figure of 1700 archived lines will rise markedly. The EMMA database gathers and curates extensive data on each line and presents it through a user-friendly website. A BioMart interface allows advanced searching including integrated querying with other resources e.g. Ensembl. Other resources are able to display EMMA data by accessing our Distributed Annotation System server. EMMA database access is publicly available at http://www.emmanet.org.
INTRODUCTION
The laboratory mouse has emerged as the major mammalian model for studying human genetic and multi-factorial diseases. Numerous mouse mutants have been produced and, more recently, technological improvements have allowed mouse mutants for virtually any gene to be produced by gene-specific approaches (knock-outs, knock-ins and conditional mutagenesis). Random approaches such as large scale, genome-wide ENU mutagenesis and gene trapping have also expanded the current repertoire of available mutants. Using these mouse mutants, researchers are able to decipher molecular disease and potentially develop new diagnostic, prognostic and therapeutic approaches.
The International Knockout Mouse Consortium [IKMC (http://www.knockoutmouse.org); (1,2)] is made up of four major projects (EUCOMM (http://www.eucomm.org) in Europe, KOMP (http://www.nih.gov/science/models/mouse/knockout/) and TIGM (http://www.tigm.org) in the USA and NorCOMM (http://www.norcomm.org) in Canada, and is in the process of producing mutations in ES cells for all known protein coding genes. A number of mouse mutant lines have already been produced from these resources. In particular, some 650 mouse lines are being produced and phenotyped in high-throughput screens as part of the EUCOMM and EUMODIC projects (http://www.eumodic.org), the results of which will be presented in the Europhenome resource (3). To take this process to the next level, the International Mouse Phenotyping Consortium (IMPC) has recently been formed with a remit to raise the funding for and to coordinate the production of mouse mutants for each of the IKMC mutations, along with high throughput phenotyping of these mice resulting in the first complete catalogue of mammalian gene function (see Appendix 6 of the PRIME final report: http://www.prime-eu.org/PRIME final report.pdf).
Archiving and distribution of the products of these various projects is a vital activity, alongside the capture of data describing in detail the genotype and phenotype characteristics of the mutants. The costs for a typical academic researcher to regenerate from scratch one of these knock-out (KO) lines has been estimated at €25–30 k and would take at least 9 months. Regenerating the mouse lines is an obvious waste of public funds for science as well as laboratory mice from an animal welfare aspect.
Since no single archiving facility can retain all of these mutant mouse strains it is essential that all mutants that have been created are held in centrally organised repositories, from which mutant mice can readily be made available to interested investigators (4,5). The European Mouse Mutant Archive [(EMMA); (6)] is a leading international network infrastructure for archiving and provision of mouse mutant strains for the whole of Europe and worldwide. To provide the best possible service to the international scientific community there is a requirement for coordination of archiving and distribution of the valuable genetically defined mice and ES cells in line with global research demand. The Federation of International Mouse Resources [(FIMRe); (7)], of which EMMA is a founding member and the European component, was initiated in response to this need for coordination.
As well as coordination of archiving, there is a requirement for a common portal that allows searching of all publicly available mice, including those not from FIMRe partners, followed by redirection to individual repositories for more detailed information and the possibility to order material. The International Mouse Strain Resource [IMSR (http://www.findmice.org); (8)] has been developed to fulfill this need and over the last few years, EMMA has become one of the largest mouse network repositories worldwide and a major contributor to IMSR.
EMMA also has a special role in the archiving and distribution of mouse mutants as it is one of four repositories handling the mouse resources produced by the IKMC initiative (EMMA archiving and distributing the mutant mice arising out of the EUCOMM project, the KOMP repository (http://www.komp.org) handling KOMP products, the Canadian Mouse Mutant Repository [CMMR (http://www.cmmr.ca); (9)] handling the NorCOMM resources and TIGM handling its own products. Eventually, these four resources will provide access to data and material covering the complete, functional characterised, proteome of the mouse, providing an unprecedented resource for bench scientists studying all aspects of the mammalian genome including human disease.
The EMMA resource database described in this paper provides up to date information about the archiving status of mice and describes the genetic and phenotypic properties of all the mutant strains that EMMA stocks. The EMMA database has two main benefits to the research community: (i) scientists with particular gene or genes of interest can discover if any mouse lines exist with mutations in these gene(s) and what the observed phenotype changes were, which may provide clues to the gene's role, and (ii) it allows scientists to order existing mouse mutants for further research and generation of data of interest to other researchers. As well as providing user-friendly searching and browsing of the database, the EMMA website is the link to the scientific community and facilitates the submission of mice to the EMMA and requests of mice from EMMA, as well as expressing interest in strains currently undergoing archiving. The data recorded for each strain is a combination of data entered by the original submitting scientist as well as subsequent curation to correct and add extra value to the database. Although the full record is only available through the EMMA database, summary data is exchanged with our partners in IKMC and the IMSR to ensure that researchers using the portals available at their sites see descriptions of EMMA lines, along with links back to the original record in EMMA and the option to order biological material. In addition, EMMA utilises the BioMart data management system (10,11) and the Distributed Annotation System [DAS; (12)] to allow distributed, integrated querying with other resources such as the Ensembl genome browser (13).
DATA COLLECTION AND CURATION
The EMMA website is used to advertise the goals of the project and encourage interested parties to submit mouse mutant lines of widespread use to the scientific research community as a disease model or other research tool. The submission process is handled automatically by the website and collects extensive data through a web form and stores this directly in the EMMA database. Data collected at this stage includes:
Contact details for the strain producer.
Strain name, affected gene(s) and mutant allele(s).
Genetic background of the original mutation and current background.
Genetic and phenotype descriptions of the line.
Bibliographic data on the line.
Whether the mouse models a human disease and an OMIM ID if appropriate.
Whether the strain is immunocompromised.
Whether homozygous mice are viable and fertile and if homozygous mating are required.
Additional optional data collected includes:
Affected chromosome, dominance pattern and ES cell line(s) used for targeted mutants.
Name and description for chromosome anomaly lines.
Mutagen used for induced mutant lines.
Promoter, founder line number and plasmid/construct name(s) for transgenic lines.
Breeding history of the line.
Current health status of the line and specific information for animal husbandry such as diet used.
How to characterise the line by genotyping, phenotyping or other methods e.g. coat colour.
Research areas the mouse is useful for, and whether it is a research tool such as a Cre-recombinase expressing line.
Extensive curation takes place to correct and augment the initial submission data. To facilitate input of correct data by submitting users, specific tools have been incorporated into the submission form, for searching and selecting approved gene, allele, background names, symbols and identifiers (from the Mouse Genome Database (MGD) developed by the Mouse Genome Informatics (MGI; http://www.informatics.jax.org) group (14). Similar tools for searching and selecting PubMed bibliographic references and identifiers have also been implemented. However, there is still a requirement for manual correction of submitted data using our curation interfaces.
The curation is based on the application of international rules and standards for the initial assignment and periodic review and update of the strain and mutation nomenclature, as defined by the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/mgihome/nomen). These approved definitions make use of control vocabularies for gene, allele, background names and symbols. Specific automated routines and associated manual curation procedures have been defined and implemented, in particular, for:
Assigning to each submitted strain record a unique EMMA identification (ID) as the primary attribute for internal strain identification and retrieval and cross-reference with connected databases such as IMSR.
Checking that the submitted records of mutant genes or expressed transgenes (and corresponding alleles), carried by the deposited strains, have assigned the correct names, symbols and identifiers, and mutation classification (as defined by MGI) according to the associated bibliographic references.
Proposing new mutant gene and allele names, symbols and identifiers for publication in the MGD database, according to the associated bibliographic references or personal communication with submitting scientists.
Checking that the submitted backgrounds of deposited strains have approved names and symbols assigned.
Inserting a preliminary strain designation for each newly submitted strain, including the assigned strain background name and the MGI allele symbol, and associating it with the corresponding EMMA strain ID.
Reviewing and approving the preliminary strain designations, in collaboration with the curation group at IMSR.
Periodically reviewing and updating of current strain designations, according to variations of MGI gene and allele's names and symbols.
Automated correction and population of bibliographic data using the submitted PubMed IDs and the CiteXplore web service (http://www.ebi.ac.uk/citexplore/).
Archiving of submitted mice is handled by one of the EMMA mouse archiving partners (CNR Instituto di Biologia Cellulare in Monterotondo, Italy; the CNRS Centre de Distribution de Typage et d’Archivage Animale in Orleans, France; the MRC Mammalian Genetics Unit in Harwell, UK; the Karolinska Institute in Stockholm, Sweden; the Helmholtz Zentrum München in Munich, Germany; the Wellcome Trust Sanger Institute in Hinxton; the Institut Clinique de la Souris in Strasbourg and the CNB-CSIC, Centro Nacional de Biotecnologia in Madrid). The archiving process involves genotype and/or phenotype verification of the mouse, followed by test freezing of either sperm or embryos and then checking the stock can be reconstituted from this frozen stock. Several strains are in particularly high demand as they represent extremely interesting disease models or valuable Cre-expressing lines and these are kept as live stocks facilitating a fast delivery to the customers. The EMMA lines are supplied to the research community for research purposes only and there is no charge for the cryopreservation service. Archiving of mice produced by the EUCOMM mouse production centres follows the same procedure except the initial import of data describing these lines is automated from the EUCOMM database. The EMMA database is used internally by the EMMA partners to track each mutant strain through the archiving process. For example, the status of the strain in the archiving pipeline, which centre is archiving the strain, the funding source for this archiving, which material is currently in stock and available to order is all stored in the database. EMMA archiving centres record this data using internal interfaces implemented using Java Spring and Hibernate technologies.
Requests for EMMA mice are also submitted via the EMMA website and recorded in the EMMA database. The archiving centres again track the whole process of distributing the requested mice using the database and the same internal Java interfaces.
EMMA now contains over 1700 submitted strains from 19 countries including around 50 lines from the USA, Canada and Australia. In the coming 5 years, it is predicted that there will be a tripling of the mouse lines held, largely as a result of the IKMC initiative. To date EMMA has sent out 1245 lines to requesting scientists worldwide. Although nearly 58% of the requests for mutant mouse lines were from European scientists, about one-third come from the USA and Canada and requests from Asia are steadily increasing. So far, EMMA has shipped mice to scientists from more than 500 different institutions located in 39 countries. Considering the estimated cost of generating these lines from scratch the existence of the EMMA resource has saved the worldwide community ∼€37 M and 934 years of laboratory effort.
QUERYING THE EMMA DATABASE
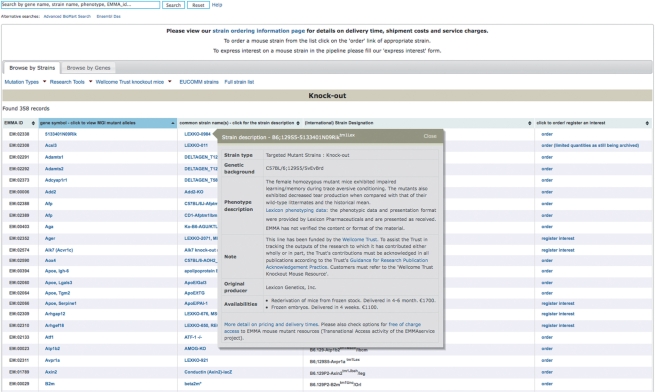
The EMMA database can be searched using a user-friendly query interface (Figure 1). This search takes full/partial case-insensitive terms and searches against the official MGI gene symbols e.g. Otog, the official IMSR designated strain name e.g. B6.129S2-Otogtm1Prs/Orl, the common strain name e.g. OtogC57BL/6J, the phenotype description e.g. auditory functions or EMMA IDs e.g. EM:01820. EMMA lines are also browsable by the affected gene, mutant type (e.g. Targeted Knock-out, Targeted Knock-in), particular research tools (e.g. Cre-expressing lines) or mice produced by large projects (e.g. EUCOMM lines). Results of searches or browsing are presented in a table, sortable by any of the columns, which lists the EMMA ID, gene affected (with hyperlinks back to MGI pages describing the particular gene and mutant alleles in detail), common strain name, approved international name and a link to either order the line or express interest in ordering lines that are in the process of being archived. The latter option triggers an automated process, in which the particular archiving centre is informed that there is a priority for this line and when it becomes available further automated emails inform the original scientist that they can go ahead and complete the ordering process.
Figure 1.
Browsing and searching for mouse lines in EMMA. Relevant strains can be identified by either (i) typing case-insensitive, full/partial terms in the top text field which searches against the affected gene symbols and name, approved international designated and common strain names, phenotype description and EMMA ID, or (ii) browsing through a complete list of lines or partial lists categorised by the affected gene(s), mutant type (targeted, gene trap, transgenic, induced, chromosomal anomalies or spontaneous), research tool [Cre recombinase expressing strains, lines for tetracycline (Tet)-regulated gene expression systems], strains provided by the Wellcome Trust Knockout Mouse Resource and finally strains produced out of the EUCOMM programme. Results are presented as a table of the EMMA ID, affected gene, common and approved international strain names alongside links to order or register interest in ordering a line when it becomes available. Clicking on any of the common strain names pops up a description of the strain.
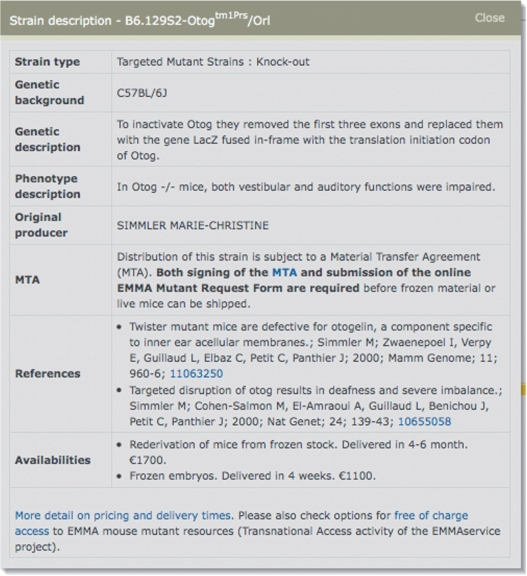
Clicking on any of the strain names pops up a strain description (Figure 2) including the mutation type, genetic background it is currently maintained on, genetic and phenotype descriptions if known, the original producer, literature references, the genotyping or phenotyping protocol needed to confirm the mutation, what material is available along with delivery times and costs and a link for downloading associated Material Transfer Agreement (MTA) documentation, if applicable.
Figure 2.
EMMA strain descriptions. Data presented includes the mutation type, genetic background it is currently maintained on, brief genetic and phenotype descriptions if known, the original producer, literature references, the genotyping protocol needed to confirm the mutation, what material is available along with delivery times and costs and a link for downloading associated MTA documentation, if applicable.
INTEGRATION WITH OTHER RESOURCES
As described earlier, a subset of data on each of the EMMA curated lines are sent weekly to the IMSR, allowing users searching this common catalogue of mutant lines to be redirected to our site for more detailed data and the ability to order the line. The MGD database provides extensive descriptions of known mutant alleles and EMMA links to the MGD pages, wherever possible as the definitive source for this data.
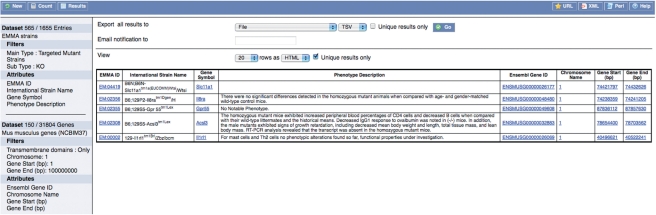
As well as our simple search box, we also provide an advanced BioMart query interface, which is linked from the main search page (Figure 3). The BioMart interface queries a denormalised snapshot of the EMMA database that is updated nightly. Queries can involve complex combinations of query terms including the affected gene symbols and MGI IDs, common and official strain names, EMMA IDs, mutant type, original and maintained genetic backgrounds and the type of material available (frozen embryos, sperm or ovaries, live mice on shelf or mice rederived from frozen stock). The results are fully configurable, allowing any combination of the fields presented in the standard EMMA search results and strain descriptions to be displayed, as well as extra data such as whether the mutant is viable and fertile when homozygous and whether it is required to keep it homozygous, whether the line is immunocompromised, if it represents a human model, the breeding history and for targeted mutants known dominance and ES cell line used, and for transgenics the promoter and plasmid construct used. The results can be previewed and exported in a number of formats such as HTML, Tab/Comma-separated text or Excel. However, the real benefit of BioMart comes from the ability to perform integrated querying with BioMarts deployed on other resources, which share a common identifier such as MGI or Ensembl IDs. For example, in Figure 3a BioMart query has identified all lines held in EMMA that have an affected gene annotated by Ensembl as being located on the first 100 Mbp of chromosome 1 and having a transmembrane protein domain.
Figure 3.
The EMMA BioMart interface. This interface allows advanced querying of the EMMA database as well as distributed and integrated querying with the Ensembl resource. In this example EMMA targeted knock-out lines are identified that have affected genes annotated by Ensembl as being located within the first 100 Mbp of chromosome 1 and containing a transmembrane domain in their protein products. The results table is fully configurable from within the interface and here shows the strain name, EMMA ID (hyperlinked back to the strain description at EMMA), gene symbol and phenotype description from the EMMA BioMart and the Ensembl Gene ID, chromosome, start and end from the Ensembl BioMart located at the Wellcome Trust Sanger Institute, Hinxton, UK.
A new portal is currently being developed for the IKMC initiative by the International-Data Coordination Center (I-DCC; http://www.i-dcc.org). This will be released late 2009 and will display the status of all genes in the mutagenesis pipeline along with available products and data for the mutant ES cells and mouse lines. The portal will utilise a number of BioMarts developed for the IKMC component mutagenesis pipelines and repositories, as well as for other useful resources such as the GXD (15) and Eurexpress (http://www.eurexpress.org) gene expression databases, and the Europhenome phenotyping resource. The EMMA BioMart will form an integral component of this IKMC portal and in addition allow a wider variety of integrated queries from our EMMA BioMart server.
Another type of data integration is provided by our Distributed Annotation System (DAS) server (www.emmanet.org/das). This serves up summary level data for each EMMA line, allowing the display of EMMA strains on DAS clients such as the Ensembl genome browser. For example by browsing to http://www.ensembl.org/Mus_musculus/Gene/ExternalData/EMMA?g=ENSMUSG00000055694 and clicking on the ‘Configure this page’ option and selecting EMMA it is possible to view any EMMA lines that exist for this gene (Gdf1). The EMMA ID, affected gene symbol, name and link to curated data at MGI is given along with the mutation type, phenotype summary and a link to the strain description at EMMA.
CONCLUSION AND FUTURE DIRECTION
The number of mutant mouse lines submitted to EMMA as well as the number of requests for these mutants is likely to increase significantly in the near future. This is due to the large scale and systematic efforts of the IKMC to perform saturation mutagenesis of the mouse genome using gene targeting and gene trapping approaches. As well as continuing to expand the number of lines curated and distributed by the EMMA resource, collaboration with international efforts to present all available mutants worldwide is going to become ever critical as the IKMC and eventually the IMPC initiatives continue to produce and characterise mutants. Data exchange with IMSR will continue to provide a common access site and EMMA will collaborate extensively with the I-DCC to provide a central portal to the data and products produced by the IKMC. There will be a particular focus on utilising the phenotyping data arising out of these programmes to allow searching for mouse models using precise phenotype queries structured using the Mammalian Phenotype (MP) ontology (16).
The EMMA project is currently funded until 2013, but obviously long term, stable funding for the data storage and mouse archiving that EMMA performs will be critical to capture and maintain the products emerging from the IKMC and IMPC programmes. This is a recognised issue and the European Commission is currently funding a number of projects under the ESFRI Roadmap with the goal of identifying sources of long term funding for key scientific activities. Infrafrontier (http://www.infrafrontier.eu) is one of these projects and is tasked with securing such funding for archiving and phenotyping of mouse mutants. Infrafrontier has already decided that the archiving aspect will be taken care of by a major upgrade to the EMMA project. Hence, it is highly likely that EMMA will continue providing this valuable service to the worldwide scientific community for many years to come.
FUNDING
European Commission FP6 Infrastructure Programme [grant no. 506455]. Funding for open access charge: European Commission FP7.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank the members of the EMMA Technical Working Group, Board of Participating Directors and EMMA archive centres who coordinate and carry out the hard task of archiving all the mouse lines.
REFERENCES
- 1.Collins FS, Rossant J, Wurst W, International Mouse Knockout Consortium A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 2.Collins FS, Finnell RH, Rossant J, Wurst W. A new partner for the international knockout mouse consortium. Cell. 2007;129:235. doi: 10.1016/j.cell.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Mallon AM, Blake A, Hancock JM. EuroPhenome and EMPReSS: online mouse phenotyping resource. Nucleic Acids Res. 2008;36:D715–D718. doi: 10.1093/nar/gkm728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbott A. Full house. Nature. 2002;417:785–786. doi: 10.1038/417785a. [DOI] [PubMed] [Google Scholar]
- 5.Marschall S, Hrabé de Angelis M. Cryopreservation of mouse spermatozoa – double your mouse space. Trends Genet. 1999;15:128–131. doi: 10.1016/s0168-9525(99)01715-1. [DOI] [PubMed] [Google Scholar]
- 6.Hagn M, Marschall S, Hrabé de Angelis M. EMMA-The European mouse mutant archive. Briefings in Func. Genomics and Proteomics. 2007;6:186–192. doi: 10.1093/bfgp/elm018. [DOI] [PubMed] [Google Scholar]
- 7.Davisson M, FIMRe Board of Directors FIMRe: Federation of International Mouse Resources: Global Networking of Resource Centres. Mamm. Genome. 2006;17:363–364. doi: 10.1007/s00335-006-0001-2. [DOI] [PubMed] [Google Scholar]
- 8.Eppig JT, Strivens M. Finding a mouse: the International Mouse Strain Resource (IMSR) Trends Genet. 1999;15:81–82. doi: 10.1016/s0168-9525(98)01665-5. [DOI] [PubMed] [Google Scholar]
- 9.McKerlie C, Ayearst R, Fleming C, Liu L, Ottaviani P, Yildiz C. The Canadian Mouse Mutant Repository: a germ cell, embryo, and tissue biorepository for functional annotation of the genome. Cell Pres. Tech. 2004;2:298–339. [Google Scholar]
- 10.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart-biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart Central Portal–unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkinson AM, Albrecht M, Birney E, Blankenburg H, Down T, Finn RD, Hermjakob H, Hubbard TJ, Jimenez RC, Jones P, et al. Integrating biological data – the Distributed Annotation System. BMC Bioinformatics. 2008;9(Suppl 8):S3. doi: 10.1186/1471-2105-9-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. Mouse Genome Database Group. The mouse genome database (MGD): new features facilitating a model system. Nucleic Acids Res. 2007;35:D630–D637. doi: 10.1093/nar/gkl940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith CM, Finger JH, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2007 update. Nucleic Acids Res. 2007;35:D618–D623. doi: 10.1093/nar/gkl1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2004;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]