Abstract
The Network of Cancer Genes (NCG) collects and integrates data on 736 human genes that are mutated in various types of cancer. For each gene, NCG provides information on duplicability, orthology, evolutionary appearance and topological properties of the encoded protein in a comprehensive version of the human protein-protein interaction network. NCG also stores information on all primary interactors of cancer proteins, thus providing a complete overview of 5357 proteins that constitute direct and indirect determinants of human cancer. With the constant delivery of results from the mutational screenings of cancer genomes, NCG represents a versatile resource for retrieving detailed information on particular cancer genes, as well as for identifying common properties of precompiled lists of cancer genes. NCG is freely available at: http://bio.ifom-ieo-campus.it/ncg.
INTRODUCTION
Cancer is a genetic disease caused by the accumulation of deleterious modifications within the genome of somatic cells (1). During tumorigenesis, genomic instability leads to the progressive acquisition of silent (‘passenger’) and selected (‘driver’) mutations (2). The latter provide cancer cells with selective growth advantages that initiate clonal expansion (3). The Cancer Genome Project (CGP) has the ambitious goal of identifying all genes that are implicated in the development of cancer (4). The Cancer Gene Census (CGC) is a part of CGP and collects information on more than 370 genes whose mutations are causally related to cancer (5). Recently, high-throughput mutational screenings of several cancer types have been promoted with the aim of identifying mutated genes, without any hypothesis-driven bias. So far, four of these high-throughput experiments have been delivered. Overall, they identified 380 Candidate Cancer Genes (CAN-genes) that are mutated in breast, colorectal, pancreatic cancers and glioblastoma (6–8). Furthermore, the pilot experiment from the Tumor Sequencing Project identified 26 genes (TSP-genes) mutated in lung adenocarcinoma (9). Altogether, these studies revealed that the number of cancer genes is surprisingly high and they are functionally more heterogeneous than previously thought. Despite this functional heterogeneity, cancer genes tend to share ‘systems-level properties’ (10), such as higher connectivity and lower duplicability when compared to the rest of human genes (11–13). The presence of shared properties, which are not strictly dependent on the gene function, indicates that cancer genes are fragile components of the human gene repertoire.
A number of databases have been set up over the years to collect and organize several types of information related to cancer, such as somatic mutations of cancer genes (14), experimental evidence for their involvement in cancer (15,16) or modifications in gene expression levels (17,18). Other databases are specialized on particular types of cancer (19,20), on single genes (21,22) or on specific genomic modifications (23,24). None of the available resources, however, focuses on properties of cancer genes that are not strictly dependent on their function, but that could help in interpreting cancer as a ‘systems disease’. Here, we present the Network of Cancer Genes (NCG, http://bio.ifom-ieo-campus.it/ncg), a database that stores information on systems-level properties of a comprehensive dataset of more than 730 cancer genes. The collected features are duplicability, evolutionary appearance and topological properties in the human protein–protein interaction network. Protein interactions have been successfully used to infer functional links between proteins (25). In NCG, they are used to understand how the topological properties of the cancer proteins inside the protein–protein interaction network influence their role in cancer. NCG can be used to retrieve information on specific cancer genes, as well as to identify groups of cancer genes with identical properties, thus providing a flexible tool for investigating the complex landscape of cancer genetic determinants. In this paper, along with a general description of the features of NCG, we also provide a specific example of how NCG can be used by reporting the properties of PTEN, a tumor suppressor gene coding for a phosphatidylinositol phosphatase that is impaired in several cancer types.
MATERIALS AND METHODS
Dataset of human cancer genes
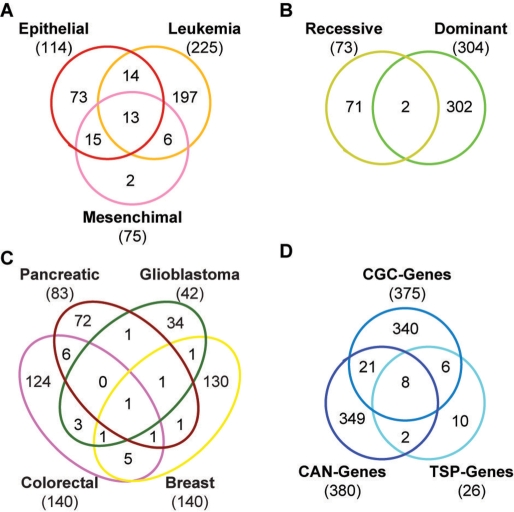
We define cancer genes as a collection of 736 genes that are mutated in different cancer types and derive from two different data sources. A total of 375 genes come from the Cancer Gene Census (CGC-genes, December 2008), a manually curated list of genes with at least two independent reports of mutations in primary tumors (5). The census provides information on the tumor type, as well as on the genetic effect of the mutation, i.e. whether the mutation is dominant or recessive (Figure 1A and B). The remaining 396 genes derive from high-throughput mutational screenings performed in glioblastoma (7), breast and colorectal (6), pancreatic (8) cancers (CAN-genes, Figure 1C) and lung adenocarcinoma (TSP-genes) (9). CAN- and TSP-genes result from the effort of massively sequencing the cancer gene repertoire (26), and provide the first unbiased mutational screenings in different cancer types. The lists of literature-curated and high-throughput derived cancer genes show poor overlap (Figure 1D), confirming the cancer-specificity of the mutational landscape (27). We gather the protein sequences associated to the 736 cancer genes from the RefSeq database [March 2009, (28)]. For eight genes no RefSeq is available and Ensembl protein sequence (29) is used instead.
Figure 1.
Cancer genes collected in NCG. Venn diagrams of the different lists of cancer genes stored in NCG. The Cancer Gene Census provides information on the cancer type (A) and on the phenotypic effect of the mutation (B). The CAN-genes reported so far refer to four cancer types (C). The overlap among the different data sources used in this study is overall very poor (D).
Gene duplicability
We define gene duplicability as in Rambaldi et al. (11). In brief, we first align the protein sequences of all human genes to the human genome reference assembly (hg18), using BLAT (30). We then retrieve the best hit of each gene, defined as the locus on the genome with the highest score in terms of coverage. By default, all genes with additional genomic matches that cover at least 60% of the query length are considered duplicable, while genes with no additional hits above this threshold are considered singleton (11). In addition to the results at the default threshold of 60%, we also provide the possibility of inspecting additional hits of the same gene covering higher or lower percentage of the original protein length. For each duplicated locus, we refer to the genome annotation provided by the UCSC Table Browser (31) to assess whether it corresponds to a known gene or instead to non-genic region.
Orthology assignment and evolutionary appearance
We derive the orthology relationships from the eggNOG database (32). Based on these relationships, we assign the evolutionary appearance of each cancer gene, defined as the deepest branch of the tree of life where an ortholog for that gene can be found. Overall, we divide the tree of life into seven main branches: Last Common Ancestor (LCA), which identifies the ancestral cellular organism, Eukaryotes, Opisthokonts, Metazoans, Vertebrates, Mammals and Primates. For example, a human gene whose orthologs are traceable in prokaryotes is considered to have appeared in the LCA, while a human gene with orthologs only in fungi and metazoans, but not in plants, is assumed to be born with Opisthokonts.
Depending on the number of paralogs of a given cancer gene at each branch, we also derive the corresponding orthology ratio, defined as the number of co-orthologs of that human gene in a given lineage. This ratio provides a useful indication of the number of intra-lineage duplications that the gene underwent during evolution. Orthology ratio can be 1 to 1 when no duplications occurred; 1 to N, indicating one-to-many relationship; N to 1, corresponding to many-to-one relationship; N to N, when multiple duplications occurred during evolution.
Protein interaction network
In order to gather the most complete representation of the human protein–protein interaction network, we integrate information from five resources: the Human Protein Reference Database (HPRD) (33), BioGRID (34), IntAct (35), the Molecular INTeraction database (MINT) (36) and the Database of Interacting Proteins (DIP) (37). We only consider primary data on interactions between human proteins, i.e. putative interactions inferred from orthology relationships are discarded. The resulting non-redundant network is composed of 68 498 interactions among 11 988 proteins, derived from 19 886 independent literature reports (Table 1). Overall, we find 4621 human proteins that interact with cancer proteins. To provide a complete view of the network of cancer proteins, NCG also allows retrieving information on the systems-level properties for all these primary interactors.
Table 1.
Integration of protein–protein interaction data
| Database | Version | Proteins | Interactions | Independent reports |
|---|---|---|---|---|
| HPRD (33) | 1 September 2007 | 8697 | 34 938 | 17 770 |
| BioGRID (34) | 1 February 2009 | 7163 | 23 588 | 8815 |
| IntAct (35) | 23 January 2009 | 7066 | 22 119 | 1374 |
| MINT (36) | 5 February 2009 | 5151 | 12 653 | 1210 |
| DIP (37) | 26 January 2009 | 1108 | 1326 | 739 |
| NCG | 21 June 2009 | 11 988 | 68 498 | 19 886 |
Data from five different sources are integrated in NCG. To derive a non-redundant version of the network, proteins are counted as number of non-redundant Entrez IDs. The number of interactions refers to non-redundant primary interactions; the independent reports refer to the number of published papers that define the interactions.
Given the poor overlap between the five data sets (Table 1), their integration allows a more complete coverage of the real interactions for each human protein. For example, the protein TP53 has a total of 408 interactions in NCG, 237 of which derive from HPRD, 214 from BioGRID, 159 from IntAct, 122 from MINT and 38 from DIP. The primary interaction network for each cancer protein is visualized using Medusa (38) and all interactors are provided with information on their duplicability, orthology, evolutionary appearance and possible involvement in cancer.
Database description
NCG is divided into four sections: (i) the gene summary table, which allows the conversion between different gene and protein identifiers, using the Entrez ID as primary key; (ii) the duplicability table, which includes all results of the BLAT alignments on the human genome; (iii) the orthology table, which stores the orthology relationships; and (iv) the network table, which includes the network properties for each protein. The data collected in NCG are stored in a MySQL database. The web interface to interrogate the database is built in Perl.
RESULTS AND DISCUSSION
Information retrieval
NCG allows retrieving information on cancer genes in three ways: (i) by using different types of identifiers, such as gene symbols, Entrez IDs (39), RefSeq (40) or Ensembl IDs (29), for specific genes or groups of genes of interest; (ii) by selecting precompiled lists of cancer genes; and (iii) by combining different criteria to analyze genes with similar duplicability, orthology and network properties. The primary output of the query is a summary table that provides links to several external databases, such as Entrez (www.ncbi.nlm.nih.gov/Entrez/), HPRD (http://www.hprd.org/), OMIM (http://www.ncbi.nlm.nih.gov/omim/) (41), RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq/) and Ensembl (http://www.ensembl.org/), as well as to detailed reports on duplicability, orthology and network properties.
Duplicability of cancer genes
In accordance with our previous report (11), at 60% coverage we find 104 duplicable cancer genes (14.1% of the total), which are associated with 336 duplicated loci. According to the available genome annotation, 44% of these additional hits correspond to known genes, 15% to more than one gene and 41% to non-genic regions. Only 22% of duplicable cancer genes duplicate in loci with no evidence of transcription, indicating that, although our measure of duplicability is based on direct genome comparison, it mostly detects transcribed paralogs.
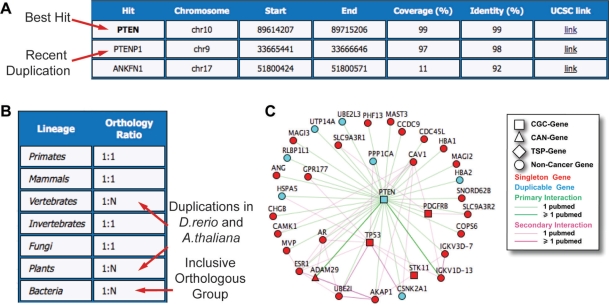
In the case of the tumor suppressor gene PTEN, we find an almost identical duplicate (97% coverage, 98% identity) corresponding to PTENP1 (Figure 2A). While the activity of PTEN as repressor of the AKT pathway is well documented (42,43), PTENP1 is known to transcribe a processed pseudogene (44,45) but the involvement in cancer has never been reported. At 10% coverage, an additional hit is found, which involves the last 50 amino acids of PTEN and matches to the intronic region between exons 3 and 4 of ANKFN1 (ankyrin-repeat and fibronectin type III domain containing 1, Figure 2A).
Figure 2.
Duplicability, orthology and network properties of the tumor suppressor gene PTEN. (A) Using the PTEN protein sequence as a query, three hits are found on the human genome. The best hit corresponds to genomic locus of PTEN, while the two additional hits account for a recent duplication transcribing for the processed pseudogene PTENP1, and to a short region of identity lying in the intron of ANKFN1, respectively. (B) The orthology ratio reflects the co-orthology relationships of human PTEN at different branching points of the tree of life. The only inparalogs of PTEN in eukaryotes are found in A. thaliana and D. rerio, indicating that this gene maintained a strict singleton status during eukaryotic evolution. (C) PTEN interacts with 35 other human proteins, four of which are cancer proteins and 22 are hubs. This makes PTEN a central node of the human protein-protein interaction network.
Orthologs and evolutionary appearance of cancer genes
NCG collects orthology information for 723 out of 736 cancer genes (98.2%), since 13 genes are not present in the eggNOG database. We find that 61% of cancer genes originated very early in evolution, because orthologs can be traced back either to LCA or to early Eukaryotes. As few as 2.5% of cancer genes appeared with Opisthokonts, 17.7% with Metazoans, 15% with Vertebrates and only the remaining 3.8% with Mammals and Primates. These results are consistent with previous reports, which assess that disease genes are overall depleted in recent genes (46).
As expected for an enzyme-coding gene, orthologs of PTEN are detectable in all branches of the tree of life, including prokaryotes, where they belong to the inclusive orthologous group of the tyrosine phosphatases. With the exception of Danio rerio and Arabidopsis thaliana, whose genomes underwent whole-genome duplications (47,48), orthologs of PTEN maintain a strict 1:1 relationship in all eukaryotic branches (Figure 2B). This suggests an early differentiation of PTEN and the maintenance of a strict singleton status during eukaryote evolution.
Network properties of cancer proteins
For each of the 579 cancer proteins with available network information (78.7% of the total), we calculate the degree, i.e. the number of interactions, the clustering coefficient, i.e. the number of interactions between primary interactors, and the betweenness, i.e. the number of shortest paths crossing the protein. These parameters return a measure of connectivity, interconnectivity and centrality, thus providing a glance of the protein topology in the network. On the basis of the network degree, we discriminate between hubs and non-hubs, where the former are defined as the top 5% most connected proteins in the network. Likewise, we identify the central nodes of the network, defined as the proteins with top 5% values of betweenness.
Overall, we find 619 human hubs, 78 of which are cancer proteins (13.4% of the total set). This result is comparable to previous reports and confirms that cancer proteins are enriched in hubs when compared to the rest of human proteins (11,12). We also observe that cancer proteins have higher betweenness than the rest of human proteins (P-value <2.2e-16, Wilcoxon test), confirming that they occupy a central position in the network.
The PTEN has overall 35 interactors, 21 of which are hubs. This, together with a high betweenness value, makes PTEN a central node that acts as a bypass between several hubs inside the human protein–protein interaction network (Figure 2C). PTEN interacts with TP53 through phosphatase-dependent and -independent mechanisms (49); it is involved in the phosphorylation of ADAM29 (50); it attenuates the activity of the tyrosine kinase receptor PDGFRB (51); and, finally, it is phosphorylated by the serine/threonine kinase STK11 (52). This confirms the tendency of cancer proteins to interact with other cancer proteins, indicating that different components of the key biological processes can contribute to tumorigenesis (11).
FUTURE PROSPECTIVE
In the coming years, we will assist to a continuous delivery of data from the Cancer Genome Project as well as from other large-scale mutational screenings of cancer genes. This massive quantity of information will require ad hoc tools for data organization and mining.
NCG represents a first attempt in the direction of a systematic analysis of cancer genes, and it will be constantly updated and expanded with the delivery of new data.
FUNDING
Associazione Italiana Ricerca sul Cancro; Fondazione Cariplo (to F.D.C.). Funding for open access charge: Associazione Italiana Ricerca sul Cancro Fondazione Cariplo.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank all members of the Ciccarelli lab for testing NCG and providing useful suggestion to improve it.
REFERENCES
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 7.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitano H. Looking beyond the details: a rise in system-oriented approaches in genetics and molecular biology. Curr. Genet. 2002;41:1–10. doi: 10.1007/s00294-002-0285-z. [DOI] [PubMed] [Google Scholar]
- 11.Rambaldi D, Giorgi FM, Capuani F, Ciliberto A, Ciccarelli FD. Low duplicability and network fragility of cancer genes. Trends Genet. 2008;24:427–430. doi: 10.1016/j.tig.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Jonsson PF, Bates PA. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22:2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez P, Huerta-Cepas J, Montaner D, Al-Shahrour F, Valls J, Gomez L, Capella G, Dopazo J, Pujana MA. Evidence for systems-level molecular mechanisms of tumorigenesis. BMC Genomics. 2007;8:185. doi: 10.1186/1471-2164-8-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr. Protoc. Hum. Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10, Unit 10 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins ME, Claremont M, Major JE, Sander C, Lash AE. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35:D721–D726. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huret JL, Dessen P, Bernheim A. Atlas of genetics and cytogenetics in oncology and haematology, year 2003. Nucleic Acids Res. 2003;31:272–274. doi: 10.1093/nar/gkg126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato K, Yamashita R, Matoba R, Monden M, Noguchi S, Takagi T, Nakai K. Cancer gene expression database (CGED): a database for gene expression profiling with accompanying clinical information of human cancer tissues. Nucleic Acids Res. 2005;33:D533–D536. doi: 10.1093/nar/gki117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elfilali A, Lair S, Verbeke C, La Rosa P, Radvanyi F, Barillot E. ITTACA: a new database for integrated tumor transcriptome array and clinical data analysis. Nucleic Acids Res. 2006;34:D613–D616. doi: 10.1093/nar/gkj022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almeida LG, Sakabe NJ, deOliveira AR, Silva MC, Mundstein AS, Cohen T, Chen YT, Chua R, Gurung S, Gnjatic S, et al. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37:D816–D819. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur M, Radovanovic A, Essack M, Schaefer U, Maqungo M, Kibler T, Schmeier S, Christoffels A, Narasimhan K, Choolani M, et al. Database for exploration of functional context of genes implicated in ovarian cancer. Nucleic Acids Res. 2009;37:D820–D823. doi: 10.1093/nar/gkn593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sedlacek Z, Kodet R, Poustka, Goetz P. A database of germline p53 mutations in cancer-prone families. Nucleic Acids Res. 1998;26:214–215. doi: 10.1093/nar/26.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cariello NF, Douglas GR, Gorelick NJ, Hart DW, Wilson JD, Soussi T. Databases and software for the analysis of mutations in the human p53 gene, human hprt gene and both the lacI and lacZ gene in transgenic rodents. Nucleic Acids Res. 1998;26:198–199. doi: 10.1093/nar/26.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knutsen T, Gobu V, Knaus R, Padilla-Nash H, Augustus M, Strausberg RL, Kirsch IR, Sirotkin K, Ried T. The interactive online SKY/M-FISH & CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes Chromosomes Cancer. 2005;44:52–64. doi: 10.1002/gcc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He X, Chang S, Zhang J, Zhao Q, Xiang H, Kusonmano K, Yang L, Sun ZS, Yang H, Wang J. MethyCancer: the database of human DNA methylation and cancer. Nucleic Acids Res. 2008;36:D836–D841. doi: 10.1093/nar/gkm730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci. Am. 2007;296:50–57. [PubMed] [Google Scholar]
- 27.Velculescu VE. Defining the blueprint of the cancer genome. Carcinogenesis. 2008;29:1087–1091. doi: 10.1093/carcin/bgn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cesareni G, Chatr-aryamontri A, Licata L, Ceol A. Searching the MINT database for protein interaction information. Curr. Protoc. Bioinform. 2008 doi: 10.1002/0471250953.bi0805s22. Chapter 8, Unit 8 5. [DOI] [PubMed] [Google Scholar]
- 37.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hooper SD, Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21:4432–4433. doi: 10.1093/bioinformatics/bti696. [DOI] [PubMed] [Google Scholar]
- 39.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's; Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 43.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 44.Dahia PL, FitzGerald MG, Zhang X, Marsh DJ, Zheng Z, Pietsch T, von Deimling A, Haluska FG, Haber DA, Eng C. A highly conserved processed PTEN pseudogene is located on chromosome band 9p21. Oncogene. 1998;16:2403–2406. doi: 10.1038/sj.onc.1201762. [DOI] [PubMed] [Google Scholar]
- 45.Kim SK, Su LK, Oh Y, Kemp BL, Hong WK, Mao L. Alterations of PTEN/MMAC1, a candidate tumor suppressor gene, and its homologue, PTH2, in small cell lung cancer cell lines. Oncogene. 1998;16:89–93. doi: 10.1038/sj.onc.1201512. [DOI] [PubMed] [Google Scholar]
- 46.Domazet-Loso T, Tautz D. An ancient evolutionary origin of genes associated with human genetic diseases. Mol. Biol. Evol. 2008;25:2699–2707. doi: 10.1093/molbev/msn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- 48.Ermolaeva MD, Wu M, Eisen JA, Salzberg SL. The age of the Arabidopsis thaliana genome duplication. Plant Mol. Biol. 2003;51:859–866. doi: 10.1023/a:1023001130337. [DOI] [PubMed] [Google Scholar]
- 49.Freeman DJ, Li AG, Wei G, Li HH, Kertesz N, Lesche R, Whale AD, Martinez-Diaz H, Rozengurt N, Cardiff RD, et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell. 2003;3:117–130. doi: 10.1016/s1535-6108(03)00021-7. [DOI] [PubMed] [Google Scholar]
- 50.Miller SJ, Lou DY, Seldin DC, Lane WS, Neel BG. Direct identification of PTEN phosphorylation sites. FEBS Lett. 2002;528:145–153. doi: 10.1016/s0014-5793(02)03274-x. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi Y, Morales FC, Kreimann EL, Georgescu MM. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. EMBO J. 2006;25:910–920. doi: 10.1038/sj.emboj.7600979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehenni H, Lin-Marq N, Buchet-Poyau K, Reymond A, Collart MA, Picard D, Antonarakis SE. LKB1 interacts with and phosphorylates PTEN: a functional link between two proteins involved in cancer predisposing syndromes. Hum. Mol. Genet. 2005;14:2209–2219. doi: 10.1093/hmg/ddi225. [DOI] [PubMed] [Google Scholar]