Abstract
MicroRNAs (miRNAs) regulate gene expression at the posttranscriptional level and are therefore important cellular components. As is true for protein-coding genes, the transcription of miRNAs is regulated by transcription factors (TFs), an important class of gene regulators that act at the transcriptional level. The correct regulation of miRNAs by TFs is critical, and increasing evidence indicates that aberrant regulation of miRNAs by TFs can cause phenotypic variations and diseases. Therefore, a TF–miRNA regulation database would be helpful for understanding the mechanisms by which TFs regulate miRNAs and understanding their contribution to diseases. In this study, we manually surveyed approximately 5000 reports in the literature and identified 243 TF–miRNA regulatory relationships, which were supported experimentally from 86 publications. We used these data to build a TF–miRNA regulatory database (TransmiR, http://cmbi.bjmu.edu.cn/transmir), which contains 82 TFs and 100 miRNAs with 243 regulatory pairs between TFs and miRNAs. In addition, we included references to the published literature (PubMed ID) information about the organism in which the relationship was found, whether the TFs and miRNAs are involved with tumors, miRNA function annotation and miRNA-associated disease annotation. TransmiR provides a user-friendly interface by which interested parties can easily retrieve TF–miRNA regulatory pairs by searching for either a miRNA or a TF.
INTRODUCTION
MicroRNAs (miRNAs) are endogenous small (∼22 nt) noncoding regulatory RNAs that typically function as negative regulators of mRNA expression at the posttranscriptional level. They act by binding to the 3′-untranslated regions (3′-UTRs) of target mRNAs through base pairing to complementary sequences. This binding results in cleavage or translation inhibition of the target mRNAs (1–3). miRNAs play critical roles in many essential biological processes, such as proliferation (4,5), metabolism (6,7), differentiation (8), development (9,10), apoptosis (7,11,12) and cellular signaling (13). Because of their biological importance, the dysfunction of specific miRNAs is associated with a variety of diseases, such as cancer and cardiovascular diseases (8,14,15).
No genes are completely independent but rather they interact with other genes. In case of miRNAs, they usually affect downstream molecules by regulating the expression of target genes. Estimates suggest that ∼1–4% of genes in the human genome encode miRNAs and that a single miRNA can regulate as many as 200 mRNAs (8). Furthermore, the expression of miRNAs can be activated or repressed by transcription factors (TFs), which therefore can serve as upstream regulators of miRNA. In recent years, many researchers have attempted to understand how miRNAs act to regulate target genes and what their roles are in various diseases. However, the study of miRNA regulation by TFs (TF–miRNA regulation) has been relatively limited. We reported previously that miRNAs and TFs may cooperate to tune gene expression (16). In addition, miRNAs and TFs can form feedback or feed-forward loops, which play critical roles in various biological processes. For example, Yamakuchi and Lowenstein (17) reported a feedback loop in which p53 induces expression of miR-34a, which in turn suppresses the expression of SIRT1 and thus increases p53 activity. Increasing evidence suggests that aberrant regulation of miRNAs by TFs can cause diseases (18). Therefore, TF–miRNA regulation is one of the most important aspects of the study of both miRNAs and TFs and is attracting the interest of increasing numbers of researchers. For this reason, a high-quality TF–miRNA regulation database will be of great help in the study of both the regulation of miRNAs by TFs and the roles of this regulation in diseases. However, such a database has not been available. To establish a database of TF–miRNA regulation, we manually curated the TF–miRNA regulatory relations that have been reported in literature and created a database that we named ‘TransmiR’. Finally, we gave a simple example for the application of TransmiR by analyzing the association between the conservation and degree of miRNAs. Although the ‘TransmiR’ database represents only the first step in this project, it should become a valuable ongoing resource for the study of TF–miRNA regulation.
DATA SOURCES AND IMPLEMENTATION
We first searched PubMed by the keywords ‘microRNA’ or ‘miRNA’ and then downloaded the search results that had been recorded before April 2009 from the National Center for Biotechnology Information (NCBI). From this search, we obtained ∼5000 papers that contained the words ‘microRNA’ or ‘miRNA’. Next, we curated the data manually and retrieved TF–miRNA regulatory pairs that related to the regulation of miRNAs by TFs. Different researchers double-checked all TF–miRNA pairs. We also noted whether a particular TF activated or repressed the expression of its miRNA partner. The database includes PubMed IDs and hyperlinks to the original PubMed articles as well as a hyperlink to NCBI (http://www.ncbi.nlm.nih.gov/) for each TF. This will enable researchers to easily access annotations such as functions, cellular components and biological processes in which the TF is involved, as well as their related disease. We annotated each miRNA according to whether it is associated with tumor and other diseases using the human microRNA disease database (HMDD, http://cmbi.bjmu.edu.cn/hmdd) (15). We also annotated the functions of each miRNA using the miRNA function database UCbase (19).
All data were organized in the ‘TransmiR’ database using SQLite, a lightweight database management system. The website is presented using Django, a Python web framework and is available at http://cmbi.bjmu.edu.cn/transmir. TransmiR provides several search options, name of the TF, miRNA ID or both.
A CASE USING TF-miRNA DATA
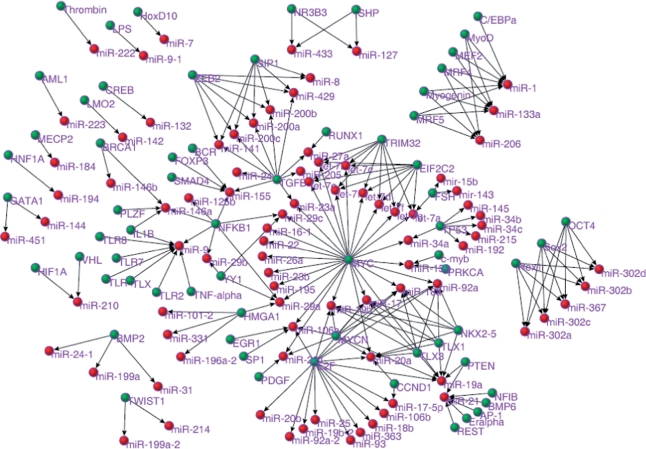
TransmiR represents a valuable resource for the study of TF–miRNA regulation and can be used to analyze various processes, such as the evolution of the interactions, expression patterns and associated diseases of miRNAs. As an example, in this study we assembled a human TF–miRNA regulatory network (Figure 1) and performed a preliminary analysis of it. As shown in Figure 1, the largest component of the network represented 67.8% of the original network nodes, which suggests that many TFs/miRNAs interact with other miRNAs/TFs. A single miRNA could be regulated by different TFs and one TF could regulate multiple miRNAs. These findings indicate that the regulatory relationships between TFs and miRNAs are complex. Both the TF and the miRNA nodes had skewed degree (the number of connections to a node) distribution. These skewed distributions suggest that most TFs regulate just a few miRNAs and, in addition, that most miRNAs are regulated by a small number of TFs. However, it also means that some hub TFs and miRNAs showed a very high number of connections, which suggests that they may play essential roles in TF–miRNA regulation. For example, MYC regulated 26 miRNAs, and miR-20a was regulated by seven TFs. Degree is a measure of node centrality in a network. Those nodes that interact with a greater number of nodes than others are normally more important in cellular functioning and could represent factors that would be more highly conserved in evolution (20). Previously, we revealed a correlation between conservation and the degree of a protein (21). However, no previous research has shown whether this pattern exists in TF–miRNA network. In order to address this issue, we investigated the correlation between conservation and degree for the miRNAs. We evaluated miRNA conservation using data on miRNA families and the method presented by Zhang et al. (22). Human miRNAs were classified into five groups according to their level of conservation: miRNAs that were present only in humans (G5), conserved in primates (G4), conserved in mammals (G3), conserved in vertebrates (G2) and those that were conserved in other more distant species (G1, the most conserved group). We classified the miRNAs in the network into two groups according to their degree: the high-degree group (degree ≥3) and the low-degree group (degree <3). We evaluated the level of conservation for the miRNAs in these two groups. As expected, we found that the miRNAs in the high-degree groups were more conserved (i.e. a greater number were in G1, P = 0.02, Fisher’s exact test) than those in the low-degree group (Table 1). This suggests that miRNAs that are regulated by a large number of TFs tend to be highly conserved during evolution.
Figure 1.
The human TF–miRNA regulatory network. Green circles represent TFs and red circles represent miRNAs.
Table 1.
Association between conservation and the degree of miRNAs
| miRNAs | Number of miRNAs in the G1 group | Number of miRNAs not in the G1 group | P-value* |
|---|---|---|---|
| High-degree group (degree ≥ 3) | 14 | 15 | 0.02 |
| Low-degree group (degree < 3) | 9 | 33 |
*P-value was calculated using Fisher’s exact test.
FUTURE EXTENSIONS
The TransmiR database represents the first step in this project and further extensions should be developed. As we described above, feedback/feed-forward loops represent two critical local interactions between TFs and miRNAs. Therefore, we plan to curate feedback/feed-forward loops between TFs and miRNAs and integrate them into TransmiR. Furthermore, we will also incorporate miRNA target data that is supported experimentally. In addition, we will classify both TFs and miRNAs into more detailed clusters according to their associations with various diseases, such as cancer or cardiovascular diseases. Finally, we will include additional annotations, such as expression patterns (23), and conservation during evolution will be included in future updates. We plan to continuously update TransmiR.
FUNDING
State Basic Research Development Program of China (No. 2007CB512100 partially). Funding for open access charge: The State Basic Research Development Program of China (No. 2007CB512100).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Drs Edmund F. and Rhoda E. Perozzi for English editing assistance.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 4.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 6.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 7.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J. Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 12.Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Cui Q, Yu Z, Purisima EO, Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol. Syst. Biol. 2006;2:46. doi: 10.1038/msb4100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ. Res. 2007;101:1225–1236. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- 15.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q. An analysis of human microRNA and disease associations. PLoS ONE. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Q, Yu Z, Pan Y, Purisima EO, Wang E. MicroRNAs preferentially target the genes with high transcriptional regulation complexity. Biochem. Biophys. Res. Commun. 2007;352:733–738. doi: 10.1016/j.bbrc.2006.11.080. [DOI] [PubMed] [Google Scholar]
- 17.Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 18.Mraz M, Pospisilova S, Malinova K, Slapak I, Mayer J. MicroRNAs in chronic lymphocytic leukemia pathogenesis and disease subtypes. Leuk. Lymphoma. 2009;50:506–509. doi: 10.1080/10428190902763517. [DOI] [PubMed] [Google Scholar]
- 19.Taccioli C, Fabbri E, Visone R, Volinia S, Calin GA, Fong LY, Gambari R, Bottoni A, Acunzo M, Hagan J, et al. UCbase & miRfunc: a database of ultraconserved sequences and microRNA function. Nucleic Acids Res. 2009;37:D41–D48. doi: 10.1093/nar/gkn702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pal C, Papp B, Lercher MJ. An integrated view of protein evolution. Nat. Rev. Genet. 2006;7:337–348. doi: 10.1038/nrg1838. [DOI] [PubMed] [Google Scholar]
- 21.Cui Q, Purisima EO, Wang E. Protein evolution on a human signaling network. BMC Syst. Biol. 2009;3:21. doi: 10.1186/1752-0509-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q, Lu M, Cui Q. 2008. SNP analysis reveals an evolutionary acceleration of the human-specific microRNAs, Nature Precedings. http://hdl.handle.net/10101/npre.2008.2127.1. [Google Scholar]
- 23.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]