Abstract
Cell division involves a complex series of events orchestrated by thousands of molecules. To study this process, researchers have employed mRNA expression profiling of synchronously growing cell cultures progressing through the cell cycle. These experiments, which have been carried out in several organisms, are not easy to access, combine and evaluate. Complicating factors include variation in interdivision time between experiments and differences in relative duration of each cell-cycle phase across organisms. To address these problems, we created Cyclebase, an online resource of cell-cycle-related experiments. This database provides an easy-to-use web interface that facilitates visualization and download of genome-wide cell-cycle data and analysis results. Data from different experiments are normalized to a common timescale and are complimented with key cell-cycle information and derived analysis results. In Cyclebase version 2.0, we have updated the entire database to reflect changes to genome annotations, included information on cyclin-dependent kinase (CDK) substrates, predicted degradation signals and loss-of-function phenotypes from genome-wide screens. The web interface has been improved and provides a single, gene-centric graph summarizing the available cell-cycle experiments. Finally, key information and links to orthologous and paralogous genes are now included to further facilitate comparison of cell-cycle regulation across species. Cyclebase version 2.0 is available at http://www.cyclebase.org.
INTRODUCTION
The process by which cells replicate and pass on their genetic information, termed the cell cycle, is fundamental to life and has been intensely studied in the biological sciences. The past decade has witnessed an explosion in data derived from cell-cycle specific and other high-throughput experiments. These data include mRNA expression profiling using microarrays (1–9), overexpression (10,11) and knock-down studies (12), prediction of degradation signals (13), and systematic determination of kinase substrates (14–16). Of particular interest are the mRNA profiling experiments, which are performed on samples aliquoted from synchronously growing cells progressing through the cell cycle. These studies provide a wealth of transcriptome data during the division process, which can be analyzed to deduce the subset of genes that are subjected to transcriptional regulation during the cell cycle. Gathering, comparing and analyzing such a vast amount of data require a significant effort.
In order to address the problems mentioned above, we developed Cyclebase (17), a web resource of cell-cycle microarray data sets and derived analysis results. The database was filled with over 20 time-series microarray experiments. In order to remove experimental condition differences and variation in the speed with which cells progress through the cell cycle, experimental data from each study were first normalized to a common time scale. Data from multiple studies were then plotted on a single chart for each gene. This intuitive visual representation, which depicts hundreds of experimental measurements in a single image, allows researchers to easily compare expression profiles across studies and gage the reproducibility of the experimental data. Each graph was supplemented by results from state-of-the-art analyses, including measures for periodicity, magnitude of regulation and the point in the division process when the transcription level is highest.
The first version of Cyclebase made it possible to easily assess transcriptional regulation of individual genes in single organisms. However, within the cell-cycle community there is a need for comparing both conservation of transcriptional regulation across species as well as assessing additional cell-cycle relevant information. To address these needs, we have expanded the functionality of Cyclebase, and further updated the database to account for changes in genomic annotations.
CYCLEBASE VERSION 2.0
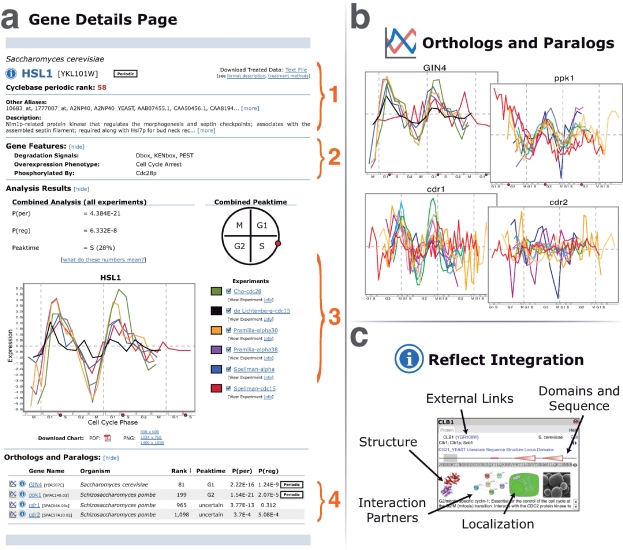
In order to provide easier access to more information about each genes’ role in the cell cycle, we have performed a major update of Cyclebase. The Gene Details page, which is the centerpiece of the web site, contains many of these updates (Figure 1). This section highlights the major additions and changes to Cyclebase and describes its core components.
Figure 1.
(a) General overview of the Cyclebase Gene Details page. (a1) Header information displays gene name, Cyclebase periodic ranking, aliases, description and links to download raw data. (a2) Annotations provide information about predicted degradation signals, kinases that phosphorylate the protein and results of overexpression and knock-down experiments. (a3) Analysis results (P-value for periodicity, P-value for regulation and peaktime value) along with an graphic depicting the peaktime are displayed above the expression chart. This chart shows all the available experiments for a given gene, each normalized to the same time scale, allowing the x-axis to be shown as phases of the cell cycle. Researchers can download the graph in both vector graphic (PDF) and image (PNG) format for use in their own publications. (a4) Orthologous and paralogous genes are shown in the same table format as the search screen. Users quickly get an overview of the similarity across organisms and can click on each gene name to see the full Cyclebase entry. (b) Clicking the chart preview icon for any ortholog or paralog expands a chart for that gene. Multiple charts can be opened simultaneously to further aid cross and inter-species gene comparisons. (c) Clicking on most gene names or information icons throughout Cyclebase provides a Reflect pop-up, which presents a variety of information about the gene selected.
Display of orthologous and paralogous genes
The recent findings that cell-cycle regulation is only rarely conserved at the individual gene level, but appears to be conserved at higher systemic levels (13), highlight the importance of comparing transcriptional regulation across species. To facilitate such comparisons, each gene is now supplemented with a list of orthologous and paralogous genes found in Cyclebase (Figure 1a4). These assignments were taken from the eggNOG database (18). This list contains analysis results, a link to display Reflect information (19), and an icon that, when clicked on, displays a graphic of all available normalized expression profiles for the ortholog or paralog selected (see Figure 1b). Multiple expression profiles can be opened at the same time, further easing comparison between homologous genes across organisms.
Addition of cell-cycle relevant data
Transcriptional regulation is one of the several regulatory layers used to control the cell cycle. Easy access to additional data relevant to the division process helps to facilitate studies that focus on the interplay between different regulatory mechanisms. Genes in Cyclebase version 2.0 now include a variety of other data related to the cell cycle. We have included cell cycle relevant features such as lists of CDK substrates (14–16), degradation motifs (13) and phenotypic effects of knock-down (12) and overexpression (10,11) experiments. These ‘gene features’ are presented on the Gene Details page (Figure 1a2).
Ability to search using BLAST
As with the original version of Cyclebase, the web-interface still queries for genes by name, alias and description. Users can continue to browse all the genes within an organism, select example genes or enter complex queries through the Advanced Search page. In addition, Cyclebase version 2.0 introduces the ability to query for genes using either animo acid or nucleotide sequence, which can be useful when performing detailed searches, e.g. searching for specific genomic sequences in the human data derived from cDNA microarray experiments. Users can either enter the primary sequence directly into the search field or use the Advanced Search feature to input a FASTA entry. Genes are queried with both BLASTP and BLASTX, the results are combined and by default are sorted by E-value.
Update to core Cyclebase components
In addition to the more visible updates, several aspects in the underlying data structure have also changed. For example, the original version of Cyclebase was organized around microarray probesets rather than genes. Multiple probesets often target the same gene and, unfortunately, single probesets may target multiple genes (i.e. there is a many-to-many relationship). Centering the new version of Cyclebase around genes, the new interface is more intuitive and warns users when a many-to-many relationship exists for the gene/probeset they are viewing. In another major change to the backend database, we have updated all data sets to account for changes in genome annotations, which provides up-to-date lists of periodically expressed genes.
Cyclebase continues to provide full documentation of analysis methodology, frequently asked questions and information on each individual experiment. In addition, well-documented downloads are available for all analysis results and, when permission from original authors has been given, normalized expression data for each experiment. All the documentation has been updated to account for the changes introduced in Cyclebase version 2.0 and all downloads have been updated with more recent genome annotations.
PERSPECTIVES
With the new functional improvements and the updated backend, Cyclebase is well positioned to store and present other temporal cell-cycle-related data sets, e.g. protein and phospho-protein expression profiles. Although only sparsely available right now, experiments that generate these types of data are expected to become more and more common in the future. Such data will help deconvolute the complexity of cell-cycle regulation, allowing researchers to further understand how regulatory mechanisms evolve, how differentiation and the cell cycle are intimately linked and how errors in the process can lead to complicated diseases such as cancer.
FUNDING
Novo Nordisk Foundation Center for Protein Research; Villum Kann Rasmussen Foundation. Funding for open access charge: Villum Kann Rasmussen Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank Hans-Henrik Stærfeldt, Kristoffer Rapacki and Peter W. Sackett for technical help with the database and Francesca Diella for help with providing data on phosphorylation sites.
REFERENCES
- 1.Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Molecular Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenberg UD, Wernersson R, Jensen TS, Nielsen HB, Fausbøll A, Schmidt P, Hansen FB, Knudsen S, Brunak S. New weakly expressed cell cycle-regulated genes in yeast. Yeast. 2005;22:1191–1201. doi: 10.1002/yea.1302. [DOI] [PubMed] [Google Scholar]
- 3.Menges M, Hennig L, Gruissem W, Murray JAH. Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 2003;53:423–442. doi: 10.1023/B:PLAN.0000019059.56489.ca. [DOI] [PubMed] [Google Scholar]
- 4.Oliva A, Rosebrock A, Ferrezuelo F, Pyne S, Chen H, Skiena S, Futcher B, Leatherwood J. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 2005;3:e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng X, Karuturi RKM, Miller LD, Lin K, Jia Y, Kondu P, Wang L, Wong LS, Liu ET, Balasubramanian MK, et al. Identification of cell cycle-regulated genes in fission yeast. Mol. Biol. Cell. 2005;16:1026–1042. doi: 10.1091/mbc.E04-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pramila T, Wu W, Miles S, Noble WS, Breeden LL. The forkhead transcription factor hcm1 regulates chromosome segregation genes and fills the s-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rustici G, Mata J, Kivinen K, Lió P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bähler J. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- 8.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niu W, Li Z, Zhan W, Iyer VR, Marcotte EM. Mechanisms of cell cycle control revealed by a systematic and quantitative overexpression screen in S. cerevisiae. PLoS Genet. 2008;4:e1000120. doi: 10.1371/journal.pgen.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Mukherji M, Bell R, Supekova L, Wang Y, Orth AP, Batalov S, Miraglia L, Huesken D, Lange J, Martin C, et al. Genome-wide functional analysis of human cell-cycle regulators. Proc. Natl Acad. Sci USA. 2006;103:14819–14824. doi: 10.1073/pnas.0604320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen LJ, Jensen TS, Lichtenberg UD, Brunak S, Bork P. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature. 2006;443:594–597. doi: 10.1038/nature05186. [DOI] [PubMed] [Google Scholar]
- 14.Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.elm: a database of phosphorylation sites–update 2008. Nucleic Acids Res. 2008;36:D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 16.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 17.Gauthier NP, Larsen ME, Wernersson R, Lichtenberg UD, Jensen LJ, Brunak S, Jensen TS. Cyclebase.org–a comprehensive multi-organism online database of cell-cycle experiments. Nucleic Acids Res. 2008;36:D854–D859. doi: 10.1093/nar/gkm729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, Powell S, von Mering C, Doerks T, Jensen LJ, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2009;38:D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pafilis E, O’Donoghue SI, Jensen LJ, Horn H, Kuhn M, Brown NP, Schneider R. Reflect: augmented browsing for the life scientist. Nat. Biotechnol. 2009;27:508–510. doi: 10.1038/nbt0609-508. [DOI] [PubMed] [Google Scholar]