Abstract
The primary objective of most gene expression studies is the identification of one or more gene signatures; lists of genes whose transcriptional levels are uniquely associated with a specific biological phenotype. Whilst thousands of experimentally derived gene signatures are published, their potential value to the community is limited by their computational inaccessibility. Gene signatures are embedded in published article figures, tables or in supplementary materials, and are frequently presented using non-standard gene or probeset nomenclature. We present GeneSigDB (http://compbio.dfci.harvard.edu/genesigdb) a manually curated database of gene expression signatures. GeneSigDB release 1.0 focuses on cancer and stem cells gene signatures and was constructed from more than 850 publications from which we manually transcribed 575 gene signatures. Most gene signatures (n = 560) were successfully mapped to the genome to extract standardized lists of EnsEMBL gene identifiers. GeneSigDB provides the original gene signature, the standardized gene list and a fully traceable gene mapping history for each gene from the original transcribed data table through to the standardized list of genes. The GeneSigDB web portal is easy to search, allows users to compare their own gene list to those in the database, and download gene signatures in most common gene identifier formats.
INTRODUCTION
Microarray gene expression profiling and other high throughput technologies have been applied to investigate and classify thousands of biological conditions. Most studies report one or more gene signatures; lists of genes that are differentially regulated between the cellular states under study, for example in a cell or tissue type, in response to treatment or at a specific time point. The value of these experimentally derived gene signatures often extend beyond their initial publication. A range of applications have been developed to use them, including Gene Set Enrichment Analysis (GSEA) which analyzes gene expression data to look for groups of genes (or gene lists) over-represented among statistically significant genes from a particular experiment (1–3). In breast cancer, a number of experimentally derived gene expression signatures including Mammaprint and Oncotype DX have been developed into commercial diagnostic assays (4) and are being validated in large scale clinical trials (5,6). Gene signatures are analyzed and validated on new gene expression data (7,8) and novel computational methods are being developed for meta analysis of gene signatures. Finally, because published experimentally derived gene signatures are typically selected to differentiate between different classes of samples, meta-analysis of multiple gene lists may provide deeper insight into the biological mechanisms underlying a wide range of processes.
While public databases such as ArrayExpress and GEO have been developed to capture gene expression data, there is no existing resource to capture the valuable end-product of the analysis of those data—the gene lists that the analyses produce. Instead, these gene lists are often included in tables or figures embedded in publications or included as supplementary material on the journal’s or the author’s website, making them generally inaccessible to automated computational analysis. If one is able to access these lists, one often finds that the lists are reported using non-standard gene identifiers, making comparison to other lists, or often to the original data, a significant challenge. To be of maximal value, gene signatures should be available through a resource that provides gene lists in a common standard format that is computationally accessible. In addition it should provide the original gene signature table as transcribed from the publication. Reproduction of a computationally accessible original transcribed gene signature table may provide additional signature meta-data, such as information and annotation about the experimental conditions and the criteria used in generating gene lists from the data (such as t-statistics scores or other ranking information) which is useful in some gene set analyses.
There have been a number of attempts to collect experimentally derived gene signatures, however these do not generally retain the original transcribed gene signature data from the publication. The largest collections of gene signature are available in MSigDB (3). MSigDB (v2.5) provides gene signatures as annotated lists of gene symbols and have curated gene signatures from 344 publications. Curators and users can submit gene lists as a two column table, containing a gene identifier and its gene symbol. However many MSigDB gene signatures contain only gene symbols, thereby limiting their future re-annotation. The Lists of Lists-Annotated (LOLA; http://www.lola.gwu.edu) database (9) contains 47 gene lists (v1.2, October 2009) and gene list input format is limited to EntrezGene or Affymetrix probeset identifiers. SignatureDB (http://lymphochip.nih.gov/signaturedb/) (10) provides 147 published and non-published gene signatures related to haematopoietic cells. The number of cancer gene signatures in Cancer Genes (http://cbio.mskcc.org/cancergenes) (11) is 26, of which 4 are from the published literature. The lack of retention of original gene list identifiers by these resources, prevents remapping of the original signatures as better genome assemblies and annotation become available (12–14). Possibly due to the limitations of the gene signature input formats, frequently little or no information is provided on the process used to map gene signatures to the identifiers that are reported.
To address these issues and facilitate gene list meta-analysis, we have systematically collected published gene signatures from publications indexed in PubMed and mapped them to a common, standardized format, and have made these available in GeneSigDB (http://compbio.dfci.harvard.edu/genesigdb). We do not collect or re-analyze the gene expression data as this is being done by other projects including gene expression atlas (http://www.ebi.ac.uk/gxa). For the GeneSigDB initial release, we reviewed over 850 published articles and manually transcribed 575 experimental cancer gene signatures from tables, figures or Supplementary Data from 319 of those papers. Signatures were curated, annotated, and mapped to the genome, providing 560 standardized gene lists. GeneSigDB provides both the original transcribed gene signature as well as the gene signature in a standard format and we publish a mapping-trace showing how each gene identifier in the original signature was re-annotated. The GeneSigDB web portal allows users to search for gene signatures and provides tools to compare gene signatures, to convert gene lists to common gene identifiers and download gene signatures in over 30 different gene identifier formats.
COLLECTION AND CURATION OF GENE SIGNATURES
Papers likely to contain one or more gene lists were first identified using PubMed searches of the form XXX AND (‘genechip’ OR microarray OR ‘gene expression’) AND (‘gene signature’ OR ‘gene list’ OR ‘expression profile’ OR ‘Classifier’ OR ‘Predictor’) AND English [la] NOT Review [pt], where XXX represents terms relevant to the particular search being conducted, such as ‘breast cancer’ or ‘stem cells.’ A full list of these terms is given in Supplementary Table S1. GeneSigDB v1.0 is based on a search of PubMed which was performed on 15 July 2009.
Each article was downloaded and gene signatures were transcribed from the manuscript or its supplementary materials. Information about the source and contents of each gene signature (Tables 1 and 2) were captured into an Excel spreadsheet template designed to capture gene signatures and associated annotation. Gene signatures appeared in a wide variety of places within particular manuscripts, including tables and graphical or textual figures (such as hierarchical clustering heatmaps) in the primary manuscripts and in supplementary pdf, excel, or text documents. Supplementary files appeared in a variety of places, including websites maintained by journals and on authors’ personal websites. Each gene signature was given a signature identifier (SigID) PMID-X, where PMID is the PubMed identifier of the article and X is the table, figure or supplementary file number from which the gene signature was extracted, for example 18490921-Table 3 indicates the gene signature was extracted from Table 3 of the article with the PMID 18490921 (15). Gene signatures were stored as tab-delimited text files and named SigID.txt. Metadata associated with each gene signature were extracted from the Excel file and stored as an xml file, SigID-index.xml, the elements of which are summarized in Figure 1 and Tables 1 and 2. The XSD schema is available File 2 in Supplementary Data. EnsEMBL gene identifiers were used as the primary gene identifier in standardized files to allow gene signature comparisons within GeneSigDB. All gene identifiers that could be mapped (listed in Table 2) to the genome were extracted into a file named SigID-mapping.txt (Figure 1) and were searched against EnsEMBL (version 55, July 2009) using BioMart (Perl API). The search history for each gene was saved in a file called SigID-maptrace.txt. If multiple gene identifiers successfully mapped to the genome, a hierarchal ranking of identifiers was used to select the best gene match (see online documentation for full description of mapping process). Standardized gene lists were stored in a file named SigID-standardized.txt. An individual directory was created for each PMID, which stored a PDF of the source manuscript and the five files derived from the gene signatures; SigID.txt, SigID-mapping.txt, SigID-maptrace.txt, SigID-standardized.txt and SigID-index.xml (Figure 1). These files are respectively, the original gene signature, the mappable gene identifiers from SigID.txt, the mapping-trace showing how each gene was mapped, a list of EnsEMBL gene identifiers that correspond to genes in SigID.txt, and xml annotation of SigID.txt.
Table 1.
Gene Signature Metadata (GeneSigDB v1.0)
| Name | Description |
|---|---|
| PMID | PubMed identifier |
| Tissue | Name of search term set used to search PubMed. (Supplementary Table S1) |
| Organism | Species common name (human, mouse, etc) |
| Platform | Name of microarray or other experimental technique used to derive gene signature (selection from constrained list, Supplementary Data S2) |
| Platform description | Description of platform |
| Genes article | Number of genes in gene signature (as described in the text of the article) |
| SigID | Signature identifier, in the format PMID-XXX, where XXX is the gene signature table, figure or supplementary file e.g. 18490921-Table 3 |
| Sig name | Name of gene signature, in the format Tissue_AuthorYear_NumberofGenes _Description. Description is optional. e.g. Breast _Bertucci08_75genes |
| Sig description | Description of gene signature, typically extracted from table or figure legend (free text) |
| File associated | Name of tab delimited file gene signature file. Format is SigID.txt |
| URL | URL from where gene signature was downloaded |
| Column mappings | Content of each column in gene signature file (selection from constrained list in Table 2) |
Table 2.
Column mappings (GeneSigDB v1.0)
| Mapping element | Description | Mapping filea |
|---|---|---|
| Probe ID | Platform specific identifier | Yesb |
| Clone ID | IMAGE clone identifier | Yesc |
| GenBank ID | GenBank accession number | Yesc |
| UniGene ID | Unigene identifier. | Yes |
| EntrezGene ID | EntrezGene or LocusLink identifier | Yes |
| Gene symbol | HGNC official gene symbol | Yes |
| CCDS ID | Consensus Coding Sequence Database ID | Yes |
| EnsEMBL ID | EnsEMBL gene ID | Yes |
| RefSeq ID | RefSeq gene identifier | Yes |
| Protein ID | Protein sequence ID, SwissProt, UniProt | No |
| Chromosome map | Chromosomal localization data | No |
| Geneset specific factor | Factor or classifier specific to data, character | No |
| Geneset specific statistics | Fold change, Ranking of genes, T-statistics, correlation, p-value, numeric | No |
| Gene description | Description or title of gene | No |
| Other gene description | KEGG, GO terms, Keywords, etc | No |
aYes indicates these columns were extracted to SigID-mapping.txt for searching biomart.
bNot all platform Probe IDs are sequence mapped in biomart. For some common unmapped microarrays, we sequence matched the probe sequences to the genome. Others were ignored.
cGenBank EST and IMAGE clone ID sequences are not in EnsEMBL and these were mapped via Unigene (See Methods).
Table 3.
Number of articles processed and gene signatures extracted by species
| Human | Mouse | Rat | Total | |
|---|---|---|---|---|
| Publications | 263 | 39 | 8 | 308 |
| Gene signatures | 465 | 84 | 11 | 560 |
| Genes | 14 197 | 9755 | 773 | – |
| Number of platforms | 32 | 9 | 4 | 38 |
| Average genes/signature | 132 | 213 | 88 | – |
Figure 1.
Curation of gene signatures in GeneSigDB. (A) GeneSigDB hierarchical file structure SigID.txt, SigID-mapping.txt, SigID-maptrace.txt, SigID-standardized.txt and SigID-index.xml. These files are respectively, the original gene signature, the mappable gene identifiers from SigID.txt, the mapping-trace showing how each gene was mapped, a list of EnsEMBL gene identifiers that correspond to genes in SigID.txt, and xml annotation of SigID.txt. (B) An example xml gene signature annotation file.
GeneSigDB RELEASE 1.0
The initial release of GeneSigDB provides 560 curated, standardized gene signatures related to cancer of the breast, ovary, lung, colon, skin, prostate, bladder, endometrial, kidney, thyroid and gene signatures related to stem cells (Tables 3 and 4). These are principally derived from gene expression studies in human tumors and cell lines (n=465) but also include additional signatures from mouse (n = 84) and rat (n = 11) (Table 3). We have curated a number of other species but have not presently mapped these to EnsEMBL genomes.
Table 4.
Number of articles processed and gene signatures extracted by search terms (tissue)
| Search terms | Number of manuscripts (articles) |
Gene signatures |
Average of genesa | ||||
|---|---|---|---|---|---|---|---|
| PubMed hits | Downloaded and processed | Curated | Mapped | Curated | Mapped to EnsEMBL | ||
| Bladder | 64 | 56 | 10 | 10 | 18 | 18 | 115 |
| Breast | 471 | 241 | 134 | 131 | 243 | 238 | 190 |
| Colon | 95 | 54 | 20 | 20 | 35 | 35 | 84 |
| Endometrial | 15 | 15 | 5 | 4 | 9 | 8 | 16 |
| Kidney | 12 | 12 | 4 | 3 | 7 | 6 | 55 |
| Liver | 129 | 27 | 8 | 8 | 12 | 12 | 144 |
| Lung | 167 | 101 | 29 | 28 | 42 | 41 | 34 |
| Ovary | 108 | 72 | 28 | 28 | 41 | 41 | 75 |
| Prostate | 136 | 102 | 30 | 28 | 52 | 48 | 47 |
| Skin | 8 | 8 | 3 | 3 | 9 | 9 | 28 |
| Stem cell | 190 | 141 | 45 | 42 | 104 | 101 | 205 |
| Thyroid | 26 | 14 | 2 | 2 | 2 | 2 | 16 |
| Uterus | 54 | 8 | 1 | 1 | 1 | 1 | 45 |
| Total | 1475 | 851 | 319 | 308 | 575 | 560 | |
aAverage number of genes per signature which were mapped to EnsEMBL genes.
The number of gene signatures per tumor type varies considerably (Table 4). Breast cancer which has been subjected to extensive gene expression profiling resulting in a new molecular subtype categorization and commercial diagnostic signatures (4), has a high number of gene signatures (n = 238). There is also a large number of gene signatures from stem cell research (n = 101), which are divided by those reported in studies of human (n = 52) and mouse (n = 49) in GeneSigDB. But in other fields of cancer research we have fewer gene signatures (kidney n = 6, endometrial n = 8). The average number of genes per signature across all gene signatures is 81 genes. However, we see variation is the number per tissue which again may reflect the ‘maturity’ of the analysis in a particular tumor. There is a broad correlation between the average number of genes per gene signature and the number of signatures collected for that tumor.
Gene loss when mapping gene signatures
The lack of standardization in reporting gene lists, and the continued evolution of the genome sequence and its annotation cause some loss in mapping gene signatures to standard identifiers. As can be seen in Table 5, many published gene signatures do not provide probe identifiers when reporting signatures despite the fact that the primary identification of the gene lists reported relies on the array probes (or probesets) rather than the genes themselves. Authors tend to report gene names or gene symbols but rarely provide details on how the gene annotations were obtained or the version of the database that was used for mapping. The latter can be important as some databases such as UniGene ‘retire’ cluster identifiers or change the gene associated with a particular cluster and other resources that rely on the genome and its annotation can change associations as the genome sequence is refined over time. In 15 of 575 curated signatures, no mappable gene identifiers were provided, the authors publish their gene signature simply as a set of gene descriptions. In Table 6, it can be seen that the success of mapping is greatly affected by the identifier provided. One lesson that clearly emerges from this analysis is that those identifiers closest to the primary data, such as probe identifiers, have the highest rate of mapping to the EnsEMBL geneIDs that are our standard identifiers. This failure in mapping severely limits our ability to compare gene lists between studies, underscoring the need for standardization in reporting gene lists. Although in Table 6 it appears that there is a low mapping success rate for EMBL/GenBank identifiers (16% success), this is a subset of EMBL/GenBank identifiers and this low mapping rate is an artifact of the search approach we are using that will be corrected in the next release GeneSigDB.
Table 5.
Frequency of different gene identifiers in mapped gene signatures
| Identifier | Frequency all gene signatures (n = 560) | Frequency human gene signatures (n = 465) |
|---|---|---|
| Gene description | 476 | 381 |
| Gene symbol | 432 | 359 |
| Probe ID | 212 | 173 |
| GenBank ID | 128 | 109 |
| UniGene ID | 96 | 75 |
| RefSeq ID | 75 | 53 |
| EntrezGene ID | 49 | 39 |
| Clone ID | 30 | 26 |
| EnsEMBL ID | 11 | 9 |
Table 6.
Success of matching different gene signature identifiers to an EnsEMBL gene
| ID type | Species | Success (unique IDs) | Failures (unique IDs) | Percentage success |
|---|---|---|---|---|
| affy_hg_u133_ plus_2 | Human | 11085 | 942 | 92 |
| affy_u133_x3p | Human | 180 | 10 | 94 |
| affy_hg_u133a | Human | 6384 | 288 | 95 |
| affy_hg_u95av2 | Human | 613 | 23 | 96 |
| affy_hg_u95a | Human | 25 | 2 | 92 |
| affy_hugenefl | Human | 43 | 8 | 84 |
| affy_mouse430_2 | Mouse | 3816 | 195 | 95 |
| affy_moe430a | Mouse | 3409 | 159 | 95 |
| affy_mg_u74av2 | Mouse | 2156 | 317 | 87 |
| affy_mg_u74a | Mouse | 116 | 1 | 99 |
| agilent_ wholegenome | Human, mouse | 3896 | 614 | 86 |
| Entrezgene | Human | 9859 | 486 | 95 |
| refseq_dna | Human | 5586 | 577 | 90 |
| ensembl_gene_id | Human | 3120 | 486 | 86 |
| hgnc_symbol | Human | 5254 | 2908 | 64 |
| mgi_symbol | Mouse | 1247 | 788 | 61 |
| rgd_symbol | Rat | 301 | 135 | 69 |
| unigene | Human, mouse | 2131 | 1865 | 53 |
| embl (genbank) | Human | 308 | 1541 | 16 |
Overlap in genes among gene signatures
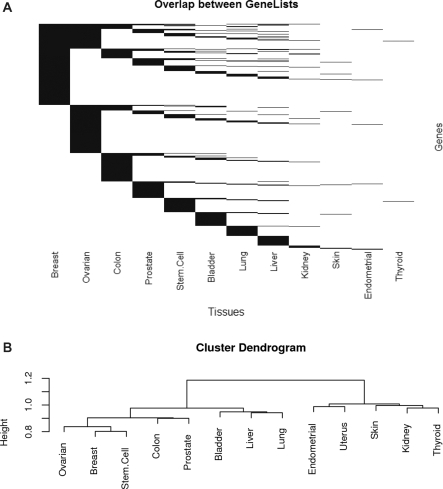
Having assembled GeneSigDB, obvious questions are which genes are most common in the various signatures that have been reported and whether there is significant overlap between reported signatures in various cancers. To analyze the overlap between gene signatures, the union of all gene lists was compared to each individual list to generate a binary matrix of presence (1) or absence (0) calls of each gene in each gene list. GeneSigDB human gene signatures (n = 465) contain 14 197 EnsEMBL genes. Histograms showing the distribution of genes in human and mouse gene signatures are provided File 3 in Supplementary Data. A large number of genes only occur in only 1 of 465 gene signatures (n = 3611), and 10 586 genes occur in 5 or fewer of the 465 gene signatures. We used a simulation approach to estimate the number of genes which might be in gene signatures by chance (described File 3 in Supplementary Data) and excluded low abundance genes. We investigated the overlap of the remaining 9920 human genes across 465 gene signatures in GeneSigDB. Figure 2 shows the overlap in gene content across all gene signatures. It can clearly be seen that related tumors such as breast and ovarian have a large overlap relative to other tumor types. This may reflect the fact that breast and ovarian cancers are known to have some common genetic components, such as mutations in the BRCA1 and BRCA2 genes. Figure 2b shows a hierarchical clustering of genes in signatures that was performed using a Sorensen’s coefficient asymmetrical measure of binary distance which gives double weight to presence and ignores absence, as we assume that presence of a gene in two lists is more informative than its absence in two gene lists. An absent gene may not be truly absent; gene expression platforms may not sample the entire genome, the probes for a particular gene may be ineffective, or the applied feature selection approach may prove sub-optimal. Since the gene x gene signature overlap matrix is sparse, scoring a double-zero between two gene lists would result in high similarity scores for many gene lists containing only a few genes. In Figure 2b, we see tumor types which are well represented in GeneSigDB cluster apart from those for which we have fewer numbers of gene signatures. However it is intriguing to observe that breast, ovarian and stem cell, colon and prostate signatures cluster, and this may reflect common etiology in these cancers.
Figure 2.
Overlap in gene signatures across tumor types. (A) Heatmap-style representation of gene overlap. Each row is a gene and each column is a tissue type. Presence of a gene is indicated by a black line, and absence is white. It can be seen that some gene maybe linked to phenotypic subclasses in many tumors, but there are generally many more tumor-specific genes, likely indicating the effect of the tissue of origin. (B) Dendrogram of hierarchical cluster analysis that was performed using a Sorensen's coefficient asymmetrical measure of binary distance and joined using Ward's minimum variance method.
We investigated if there were genes that occur frequently in many gene signatures. We observed 80 genes occur in 25 or more gene signatures. The most frequently observed genes occur in 50 of the 465 genes signatures and are MAD2LI (ENSG00000164109) and RRM2 (ENSG00000171848). However, these occur predominately in breast genes signatures (n = 42/50). We therefore examined which genes are common in many tissues types and observed 29 genes occur in 7 or more tissue types (Table 7). We performed a representational analysis to search Gene Ontology terms and KEGG pathways that are over-represented in that set. Not surprisingly, the most common functional classes found were those associated with cell cycle, consistent with the fact that cancer is a disease that disrupts normal cell cycle control (File 3 in Supplementary Data).
Table 7.
Most common genes across genes signatures from all tissue types
| EnsEMBL Gene ID | Hgnc symbol | Number tissue types | Counts of gene signatures in tissue types |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bld | Br | Co | End | Kd | Li | Lu | Ov | Pr | Sk | SC | Thy | Ut | |||
| ENSG00000113140 | SPARC | 9 | 2 | 24 | 2 | 0 | 0 | 1 | 2 | 3 | 1 | 3 | 1 | 0 | 0 |
| ENSG00000115414 | FN1 | 8 | 1 | 22 | 1 | 0 | 0 | 2 | 1 | 7 | 5 | 0 | 2 | 0 | 0 |
| ENSG00000131747 | TOP2A | 8 | 0 | 30 | 1 | 0 | 1 | 1 | 1 | 3 | 2 | 0 | 4 | 0 | 0 |
| ENSG00000134755 | DSC2 | 8 | 0 | 8 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 |
| ENSG00000151914 | DST | 8 | 0 | 15 | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 2 | 3 | 0 | 0 |
| ENSG00000157456 | CCNB2 | 8 | 0 | 27 | 1 | 0 | 1 | 0 | 2 | 2 | 1 | 0 | 2 | 1 | 0 |
| ENSG00000134057 | CCNB1 | 8 | 0 | 22 | 1 | 1 | 0 | 0 | 3 | 1 | 3 | 1 | 2 | 0 | 0 |
| ENSG00000087586 | AURKA | 8 | 0 | 33 | 1 | 2 | 1 | 0 | 0 | 1 | 1 | 1 | 4 | 0 | 0 |
| ENSG00000120992 | LYPLA1 | 8 | 1 | 6 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 2 | 0 | 0 |
| ENSG00000132646 | PCNA | 7 | 1 | 19 | 2 | 0 | 0 | 0 | 2 | 3 | 2 | 0 | 1 | 0 | 0 |
| ENSG00000169429 | IL8 | 7 | 2 | 14 | 3 | 0 | 0 | 0 | 1 | 3 | 2 | 0 | 2 | 0 | 0 |
| ENSG00000121966 | CXCR4 | 7 | 1 | 16 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 3 | 0 | 0 |
| ENSG00000139318 | DUSP6 | 7 | 0 | 11 | 1 | 0 | 0 | 2 | 1 | 1 | 1 | 0 | 2 | 0 | 0 |
| ENSG00000146674 | IGFBP3 | 7 | 0 | 8 | 2 | 0 | 0 | 1 | 1 | 4 | 3 | 0 | 1 | 0 | 0 |
| ENSG00000170312 | CDC2 | 7 | 0 | 28 | 1 | 1 | 0 | 0 | 2 | 1 | 2 | 0 | 4 | 0 | 0 |
| ENSG00000185275 | CD24L4 | 7 | 0 | 15 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 0 |
| ENSG00000164171 | ITGA2 | 7 | 0 | 5 | 1 | 0 | 1 | 1 | 0 | 1 | 2 | 0 | 2 | 0 | 0 |
| ENSG00000176890 | TYMS | 7 | 1 | 17 | 2 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 2 | 0 | 0 |
| ENSG00000044115 | CTNNA1 | 7 | 1 | 8 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 |
| ENSG00000164442 | CITED2 | 7 | 3 | 10 | 2 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 0 |
| ENSG00000003436 | TFPI | 7 | 0 | 10 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 2 | 1 | 0 | 0 |
| ENSG00000108821 | COL1A1 | 7 | 1 | 15 | 0 | 0 | 0 | 1 | 4 | 1 | 1 | 0 | 4 | 0 | 0 |
| ENSG00000111348 | ARHGDIB | 7 | 1 | 10 | 0 | 0 | 0 | 3 | 2 | 1 | 1 | 0 | 1 | 0 | 0 |
| ENSG00000196139 | AKR1C3 | 7 | 1 | 5 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 3 | 0 | 0 |
| ENSG00000204262 | COL5A2 | 7 | 1 | 13 | 0 | 0 | 0 | 0 | 2 | 2 | 1 | 2 | 1 | 0 | 0 |
| ENSG00000142871 | CYR61 | 7 | 1 | 11 | 0 | 0 | 0 | 1 | 0 | 1 | 4 | 1 | 2 | 0 | 0 |
| ENSG00000170345 | FOS | 7 | 1 | 24 | 0 | 0 | 0 | 1 | 0 | 1 | 5 | 0 | 2 | 0 | 1 |
| ENSG00000167642 | SPINT2 | 7 | 0 | 7 | 0 | 0 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 0 | 0 |
| ENSG00000175063 | UBE2C | 7 | 0 | 25 | 0 | 0 | 1 | 0 | 1 | 2 | 1 | 0 | 1 | 1 | 0 |
Tissue types are Bladder (Bld), Breast (Br), Colon (Co), Endometrial (End), Kidney (Kd), Liver (Li), Lung (Lu), Ovary (Ov), Prostate (Pr), Skin (Sk), Stem Cell (SC), Thyroid (Thy), Uterus (Ut).
THE GeneSigDB WEB INTERFACE
Querying GeneSigDB
There are two entry points into GeneSigDB, a publication-based and a gene-based search. The publication search queries articles and retrieves a list of publications and the signatures they describe. The results are based on two independent searches. The first is a full-text search of the articles indexed in GeneSigDB. This search includes the article title, authors, affiliations, abstract, introduction, methods, results, discussion and other items in the main body of the publication; the reference section is not included in this search. A second search queries only the information indexed by PubMed which includes title, author names, journal, title and abstract. The most common publication search terms would be an author name, article title, journal name or keywords. Terms can be combined using standard Boolean operators and examples are provided in the documentation online.
The gene search queries the annotation of genes within indexed signatures. One can enter either a single gene or multiple genes into the gene search. Gene search terms can be gene symbols, EnsEMBL, Entrezgene, Affymetrix, Illumina or other common microarray probe identifiers. A gene list can be entered in space or comma separated format. Wildcard searches are permitted, for example BRCA* will return BRCA1 and BRCA2 genes. Examples of both publication and gene searches are provided in the help documentation online.
GeneSigDB data views
There are three data views in GeneSigDB; a publication view, a gene signature view, and a gene view. The first is the publication view which contains information about the publication (authors, title, journal, publication date and abstract) and links to all gene signatures extracted from that publication. The second is the gene signature view which presents the gene signature metadata (described in Tables 1 and 2) and data related to the gene signature (Figure 1), including the original transcribed gene signature table and a standardized gene list of EnsEMBL identifiers and gene symbols. GeneSigDB also provides a history of how each gene was mapped to EnsEMBL. When a gene cannot be mapping to an EnsEMBL gene, this is clearly stated. The third data view is the gene view, which provides gene annotation information such as gene synonyms, description and gene identifiers of popular databases (EnsEMBL, EntrezGene, RefSeq). Where a gene signature is of non-human origin, the human orthologue of the gene is provided (where possible). A gene view lists all gene signatures in which that gene can be found.
Visualization of overlap between gene signatures
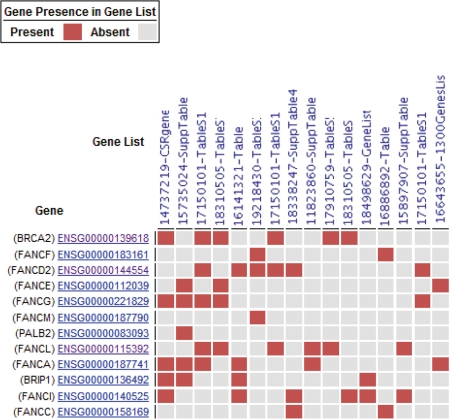
To visualize the overlap between multiple gene signatures, one can tick checkboxes selecting multiple gene signatures in several views including the publication search results, gene search results, or gene or publication entry view. These gene signatures are passed to a gene signature comparison view. This opens a gene × signature comparison matrix in which the rows are genes and the columns are signatures; the elements of the matrix are colored heatmap-style red or grey to represent presence or absence respectively. The default setting is that only genes present in two or more signatures are shown. As an example, Figure 3 shows the overlap of Fanconi anemia associated genes in GeneSigDB gene signatures. This analysis is based on a gene search with the wild card search FANC* which returned 12 human genes and 2 mouse genes. We selected the 12 human genes by ticking the checkboxes and then clicked on the compare button to visualize the overlap in these in the comparison view.
Figure 3.
Visualization of gene overlap in gene signatures. In this example, we searched for Fanconi anemia-related genes by performing a gene search for FANC* which returned 12 human genes and 2 mouse genes. This screen shows the overlap between gene signatures in which these 12 genes are present in at least 2/12. Red and grey boxes indicate presence or absence, respectively.
Downloading gene signatures
In the publication search results, gene search results or gene or publication entry view, gene signatures can be selected for download using checkboxes which are passed to a download page. There, a user can choose to download the standard gene list (EnsEMBL gene identifiers and gene symbols) or can choose to convert gene signatures into one or many commonly used identifiers, including Entrezgene, ReqSeq gene identifiers or Affymetrix, Agilent or Illumina probe identifiers. There is no limit to the number of identifiers that can be selected or to the number of gene signatures that can be downloaded concurrently. Each gene signature is provided in a separate comma separated file and if multiple gene signatures are downloaded together, these are compressed into one zip file.
Architecture of Web interface to GeneSigDB
The web interface to GeneSigDB (http://compbio.dfci.harvard.edu/genesigdb) is based on HTML, CSS, JSP, XML and Java 1.6 technologies. The application runs on an Apache Tomcat 6 web application server running on a CentOS 5 Linux server. Front-end interactivity makes use of the jQuery 1.3.2 Javascript library and server-side processing is based on the Apache Lucene 2.0 framework. GeneSigDB is not based on traditional database technology since the application does not require a high level of read/write access. Instead, the full text of articles along with numerous other curated resources are indexed and cross-referenced using the Lucene model (http://lucene.apache.org/) to produce a high-performance search system.
Comparison of GeneSigDB to other gene signature resources
GeneSigDB provides a large collection of experimentally derived gene signatures. To the best of our knowledge, only MSigDB contains more curated gene signatures. MSigDB contains 1186 curated (c2) gene signatures from 344 publications, however the overlap between MSigDB and GeneSigDB is minimal. Only signatures from 13 publications are contained in both MSigDB and GeneSigDB. Consequently GeneSigDB release 1.0 provides a large number of cancer and stem cell gene signatures that were not previously computationally accessible.
One fundamental aspect of GeneSigDB that differs from existing resources is the importance given to traceability of each gene signature. Each gene signature has a signature identifier PMID-X, where PMID is the PubMed identifier of the article and X is the table, figure or supplementary file number from which the gene signature was extracted, so that it can be easily traced to the original publication. In addition we provide a transcribed copy of the original table from the article. A fully traceable gene history for each gene from the original transcribed data table through to the standardized list of genes is also provided, including version number of all databases used in generating gene annotation. Therefore the source gene of identifiers in each standardized gene list should be unambiguous. Since original gene identifiers are stored and formatted in an annotation pipeline, GeneSigDB standardized gene lists and annotation will be updated with each release of GeneSigDB.
SUMMARY AND FUTURE DIRECTIONS
GeneSigDB fills an important need within the community—the need to standardize gene expression signatures to facilitate comparison and to allow them to be easily queried and used in other analyses. GeneSigDB release v1.0 focused on cancer and stem cell gene signatures because these together represent some of the largest sources of gene expression-based signatures. Because of the way in which these signatures were identified, we anticipate that they may capture many of the underlying processes associated with the development and progression of cancers and that their comparison may yield additional insight into the disease. We also recognize that many of these processes likely are important in other disease and non-disease phenotypes. Consequently, we plan to expand GeneSigDB to include a broader range of gene signatures, both from other disease-based studies and signatures arising from different technologies such as copy number variation arrays. The GeneSigDB web site interface will continue to improve and we intend to implement several new features that will vastly improve the gene signature comparison visualization. We are also working to expand gene signature annotation in GenSigDB to provide web links to GEO or ArrayExpress datasets (where applicable), and biological sample information on the source of each gene signature, and in future also hope to implement a controlled vocabulary to enable better searching and analysis of gene signatures.
One lesson that we have learned from GeneSigDB is that there is a pressing need for standardization of gene expression signatures. Our creation of this database grew out of a desire to do a simple computational analysis of published gene expression signatures to look for similarities between tumors arising in different organ sites. While databases such as ArrayExpress and GEO have become valuable repositories for the raw data from expression studies, the gene expression signatures that are the results of expert analysis of those data are currently not stored or reported in a systematic fashion. While GeneSigDB represents an attempt to remedy the situation, the need for extensive manual assembly and curation argues for the development of standardized reporting formats for gene signatures to facilitate their broader use and reuse.
LICENSE
The software used in constructing GeneSigDB is open source software and provided under the Artistic License. All content within GeneSigDB is provided without restriction.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: US National Institutes of Health (grant numbers R01-CA098522 and 1P50HG004233); the Dana-Farber Cancer Institute Women’s Cancer Program; and funds provided through the Dana-Farber Strategic Plan Initiative.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to acknowledge assistance from the Dana-Farber Cancer Institute Center for Cancer Computational Biology and are grateful for discussions and assistance of Ms Kristina Holton, Dr Stefan Bentink and Dr Joseph White. We thank Dr Oliver Hofmann and Prof Winston Hide for their collaborative assistance in curation of stem cell gene signatures.
REFERENCES
- 1.Oron AP, Jiang Z, Gentleman R. Gene set enrichment analysis using linear models and diagnostics. Bioinformatics. 2008;24:2586–2591. doi: 10.1093/bioinformatics/btn465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goeman JJ, Bühlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23:980–987. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- 3.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross JS. Multigene classifiers, prognostic factors, and predictors of breast cancer clinical outcome. Adv. Anat. Pathol. 2009;16:204–215. doi: 10.1097/PAP.0b013e3181a9d4bf. [DOI] [PubMed] [Google Scholar]
- 5.Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J. Clin. Oncol. 2008;26:721–728. doi: 10.1200/JCO.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 6.Cardoso F, Veer LV, Rutgers E, Loi S, Mook S, Piccart-Gebhart MJ. Clinical application of the 70-gene profile: the mindact trial. J. Clin. Oncol. 2008;26:729–735. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
- 7.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DSA, Nobel AB, van’tVeer LJ, Perou CM. Concordance among gene-expression-based predictors for breast cancer. N. Engl. J. Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 8.Chang HY, Nuyten DSA, Sneddon JB, Hastie T, Tibshirani R, Sørlie T, Dai H, He YD, van’tVeer LJ, Bartelink H, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc. Natl Acad. Sci. USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cahan P, Ahmad AM, Burke H, Fu S, Lai Y, Florea L, Dharker N, Kobrinski T, Kale P, et al. List of lists-annotated (lola): a database for annotation and comparison of published microarray gene lists. Gene. 2005;360:78–82. doi: 10.1016/j.gene.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Shaffer AL, Wright G, Yang L, Powell J, Ngo V, Lamy L, Lam LT, Davis RE, Staudt LM. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol. Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 11.Higgins ME, Claremont M, Major JE, Sander C, Lash AE. Cancergenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35:D721–D726. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari F, Bortoluzzi S, Coppe A, Sirota A, Safran M, Shmoish M, Ferrari S, Lancet D, Danieli GA, Bicciato S. Novel definition files for human GeneChips based on GeneAnnot. BMC Bioinformatics. 2007;8:446. doi: 10.1186/1471-2105-8-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, Lee JC, Salit ML, Cam MC. Transcript-based redefinition of grouped oligonucleotide probe sets using AceView: high-resolution annotation for microarrays. BMC Bioinformatics. 2007;8:108. doi: 10.1186/1471-2105-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neerincx PB, Casel P, Prickett D, Nie H, Watson M, Leunissen JA, Groenen MA, Klopp C. Comparison of three microarray probe annotation pipelines: differences in strategies and their effect on downstream analysis. BMC Proc. 2009;3(Suppl. 4):S1. doi: 10.1186/1753-6561-3-S4-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertucci F, Orsetti B, Nègre V, Finetti P, Rougé C, Ahomadegbe JC, Bibeau F, Mathieu MC, Treilleux I, Jacquemier J, et al. Lobular and ductal carcinomas of the breast have distinct genomic and expression profiles. Oncogene. 2008;27:5359–5372. doi: 10.1038/onc.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]