Abstract
The Protein Data Bank in Europe (PDBe) (http://www.ebi.ac.uk/pdbe/) is actively working with its Worldwide Protein Data Bank partners to enhance the quality and consistency of the international archive of bio-macromolecular structure data, the Protein Data Bank (PDB). PDBe also works closely with its collaborators at the European Bioinformatics Institute and the scientific community around the world to enhance its databases and services by adding curated and actively maintained derived data to the existing structural data in the PDB. We have developed a new database infrastructure based on the remediated PDB archive data and a specially designed database for storing information on interactions between proteins and bound molecules. The group has developed new services that allow users to carry out simple textual queries or more complex 3D structure-based queries. The newly designed ‘PDBeView Atlas pages’ provide an overview of an individual PDB entry in a user-friendly layout and serve as a starting point to further explore the information available in the PDBe database. PDBe’s active involvement with the X-ray crystallography, Nuclear Magnetic Resonance spectroscopy and cryo-Electron Microscopy communities have resulted in improved tools for structure deposition and analysis.
INTRODUCTION
The Protein Data Bank in Europe [PDBe; previously known as the Macromolecular Structure Database, MSD (1)], the Research Collaboratory for Structural Bioinformatics (RCSB) (2) and the BioMagResBank (BMRB) (3) in the USA, and the Protein Data Bank Japan (PDBj), collectively form the Worldwide Protein Data Bank (wwPDB) (4) organization. wwPDB is responsible for maintaining the single worldwide repository of bio-macromolecular structure data, the Protein Data Bank (PDB) (5,6). To ensure that the core archive data are represented uniformly at all the wwPDB sites, there is an active collaboration involving exchange of the core reference information (e.g. the dictionary description for ligands) as well as regular discussions to apply uniform standards for deposition and annotation. The PDBe is also active in the maintenance of the PDB archive and runs a deposition system through which new macromolecular structures can be added to the archive.
PDBe maintains collaborations with several other major bioinformatics projects hosted at the European Bioinformatics Institute (EBI) and around the world. These partnerships allow PDBe to enhance the usefulness and scope of its databases and services by adding curated and actively maintained derived data to the existing structural data in the PDB. Our partners benefit from access to a consistent and readily accessible source of macromolecular structure data. These collaborations have led to the addition of a volume of data, such as cross-references to the SCOP (7) and CATH (8) structure classification databases, the sequence-centric databases UniProt (9), InterPro (10), Pfam (11) and ProSite (12), as well as the gene ontology (GO) database (13) and the PubMed literature archive (14). This effort has resulted in the development of Structure Integration with Function, Taxonomy and Sequence (SIFTS) initiative (15) that makes the linkage data available.
PDBe is actively working with the X-ray crystallography, Nuclear Magnetic Resonance (NMR) spectroscopy and cryo-Electron Microscopy (EM) communities. To keep abreast of new developments in the NMR community, PDBe collaborates with BMRB, has participated in EU projects (eNMR and until recently Extend-NMR), and continues to contribute to the Collaborative Computational Project for the NMR community (CCPN) (16), which aims to provide a standard model, related libraries (17), reference data (18) and applications (19) for storage and handling of NMR data. PDBe’s commitment to the field of cryo-EM dates back to 2002 when the Electron Microscopy Data Bank (EMDB) was established at the EBI (20). Originally funded through the EU-funded Network of Excellence for ‘Three-Dimensional Electron Microscopy’, EMDB is now developed and operated jointly with RCSB and Baylor College of Medicine with National Institutes of Health (NIH) funding. Recently, EMDB has also received UK funding for a collaborative project with the Open Microscopy Environment developed at the University of Dundee (21).
The main objectives of the work at PDBe are as follows:
To expertly handle deposition and annotation of structural data as one of the wwPDB deposition sites. We aim for an average turn around time of one working day or less, employ skilled annotators with experience in structure determination and eventually intend to handle roughly one-third of all depositions worldwide.
To provide an integrated resource of high-quality macromolecular structures and related data. This is implemented by developing and maintaining advanced structural bioinformatics databases and services that are or even define the state of the art. They should be kept up-to-date to keep pace with the growth of the PDB archive and ideally be available on a 24/7 basis for 360+ days per year.
To maintain in-house expertise in all the major structure determination techniques (X-ray, NMR and EM) so as to be able to stay abreast of technical and methodological developments in these fields, and to work with the community on issues of mutual interest (e.g. data representation, harvesting, formats and standards or validation of structural data).
Here, we describe some results of our recent work with the NMR and EM communities as well as some new developments in the deposition and annotation system. We also describe several new databases and services based on remediated PDB data (22).
DATA DEPOSITION AND ANNOTATION
AutoDep version 4 (http://www.ebi.ac.uk/pdbe-xdep/autodep/) is a service provided by PDBe to facilitate deposition of coordinate and experimental data into the PDB. The current release of AutoDep (4.3) contains significant improvements over previous releases of AutoDep 4 (23) and the original AutoDep system, written at BNL in the 1990s. AutoDep 4 is a complete rewrite of the deposition system using Java and XML technologies. The system is highly configurable and customizable, owing to the use of XML dictionaries to define and describe all aspects of the user interface, data flow and data-storage formats. AutoDep 4.3 is also available for download and installation locally in laboratories and comes pre-packaged with all necessary dependencies to ensure straightforward installation under Linux operating systems. The advantage of a local installation of AutoDep is that this can not only be used as an internal data-archival tool but also allows depositors to complete all necessary deposition and preliminary validation checks at their leisure. Once the internal deposition is complete, the user can upload a tar package to the public server and complete online deposition in a matter of minutes.
PDBe is committed to providing value-added data to the depositor as part of the deposition process. In addition to providing the annotated structures to the depositor, we also include detailed chemical dictionary descriptions for new ligands (22), Electron Density Server (24) validation reports for X-ray entries, quaternary structure descriptions from PISA (25) and sequence and taxonomy cross-references. The structure download, chemical dictionary and quaternary structure data are also presented in an interactive 3D-Java viewer that allows real-time manipulation and visualization of the information provided. The validation tools built into AutoDep are continually improved to reduce data-entry errors at the time of deposition. The PDBe structure validation package has also been updated to provide more useful information to the depositor prior to deposition.
AutoDep supports integration between EMDB volume deposition (26) and PDB coordinate submission. Depositors of EMDB volume data have the option to deposit fitted coordinates at the PDB. To this end, extensions have been implemented to both EMDep and AutoDep that allow information from EMDep to initiate a seamless PDB coordinate deposition via AutoDep.
The latest release of AutoDep supports the uploading of NMR structures and data as a CCPN project (16). This mechanism of PDB coordinate deposition accepts a full CCPN project file, and extracts information pertinent to PDB deposition and BMRB restraint deposition automatically, thereby reducing effort and error on part of the depositor. Before submitting a CCPN project, users can select the data that is to be made public using the graphical CcpNmr ECI (Entry Completion Interface; http://www.ebi.ac.uk/pdbe/docs/pdbe_nmr_deposition/eci.html). This desktop application allows for secure editing of data prior to submission, and in the future we aim to provide pre-submission validation report pages using software written by the CCPN team. To use the new CCPN/AutoDep server on the PDBe web pages at EBI, a user simply submits the CCPN project as a compressed tar archive. Currently, the CCPN AutoDep upload page only accepts CCPN version 2 projects. Users of CCPN version 1 software can use an upgrade script provided as a web application (http://www.ccpn.ac.uk/upgrade), which will take CCPN version 1 projects and convert them to version 2. After submission, the data are curated at PDBe and the data in the original CCPN project are updated to reflect any changes in the data content. At this point, the CCPN project is converted to NMR-STAR format and the resulting file is automatically forwarded to BMRB. Subsequently, the user has the option to complete an ADIT-NMR (Auto Dep Input Tool-NMR) deposition and further curation of the NMR data at BMRB is initiated.
PDBe DATABASE
The release of remediated coordinate files by wwPDB (22) resulted in a consistent data archive and formed the basis of the development of the new database at PDBe. The new database framework was also required to ensure long-term maintainability. The new database adopts the model and the terminology of the wwPDB standard dictionary. The design is implemented in a generic form that can be used with a variety of database engines (e.g. MySQL, http://www.mysql.com, or Oracle, http://www.oracle.com). The performance and flexibility of the new database has helped in the continued efforts towards maintaining high data quality in the archive for newly deposited data. The database ensures that the core coordinate data in the archive conform to the standard reference information, e.g. ligand names, atom names and atom elements, thus ensuring data integrity across all the entries in the PDB archive. The database also implements a mechanism to provide cross-references to the NCBI taxonomy database, journals and citations, and other biological databases such as UniProt, Pfam, SCOP and CATH. The use of standard database technology has enabled efficient exchange of data with a number of other biomedical databases such as the taxonomy database maintained by the UniProt group, the PubMed Journal database as a source of reference information about journals and the UniProt database. The new database continuously reapplies a sequence cross-reference procedure to enable resynchronization between PDB and UniProt entries on the residue level as both databases evolve. PDBe continues to support the SIFTS initiative (15) and provides the SIFTS data as a consistent, high-quality and up-to-date data resource for cross-reference information between PDB and UniProt.
To support efficient querying and data retrieval, the database design needs to be flexible. In some instances, specially designed databases are required, for instance for the PDBeMotif service. The PDBeMotif database is designed to store the core coordinate data as well as information on the intermolecular and intramolecular networks of interactions. This database further contains information on small 3D structural motifs and employs the Distributed Annotation System (27) to obtain additional annotations of motifs and sequence and structure family assignments based on a number of resources in the central Distributed Annotation System registry (http://www.dasregistry.org/). These include Catalytic Site Atlas, CATH, GENE3D, MEROPs, Pfam, PIRSF, PRINTS, PROSITE, Super Family (SSF), SMART, TIGRFAM and UniProt. For efficient querying, the data from the PDB archive are split into four categories implemented as different database tables: proteins, nucleic acids, bound molecules and solvents. This separation allows for efficient storing of interaction data calculated for all pairs of atoms with a distance ≤16 Å. Data on dihedral angles, secondary structure and small 3D motifs (28–34) are kept in additional tables. Both databases are organized in data marts that are self-contained modules of data; for instance, the coordinates and related information form the core mart, whereas the sequence, taxonomy and cross-referencing information to other bioinformatics resources forms a separate self-contained mart. The two databases described above provide the basis for various advanced search systems and analysis tools designed to deliver structural data to the user community.
PDBe SEARCH SERVICES AND TOOLS
We have developed new and enhanced search systems for the PDBe database that allow users to access independent marts of the database, as well as interfaces that cover all the marts and allow queries across the whole database. PDBe also continues to develop and maintain tools for analysis of macromolecular structures. Table 1 lists all the services available at PDBe as of September, 2009.
Table 1.
Overview of services available from PDBe as of September, 2009
| Service | Description | URL |
|---|---|---|
| BIObar | Search system implemented as a toolbar application for Mozilla browsers | http://www.ebi.ac.uk/pdbe/docs/biobar.html |
| PDBeStatus | Search system to query the status of PDB entries | http://www.ebi.ac.uk/pdbe-as/pdbStatus |
| PDBeMapQuick | Quick access to cross-reference information to external databases based on PDB ID | http://www.ebi.ac.uk/pdbe-as/PDBeMapQuick/ |
| PDBeView | Text-based and advanced PDB search tool | http://www.ebi.ac.uk/pdbe-srv/view |
| PDBeLite | Search system based on the relational PDBe database | http://www.ebi.ac.uk/pdbe-srv/pdbelite |
| EMsearch | Search system for the EM Database | http://www.ebi.ac.uk/pdbe-srv/emsearch |
| PDBeChem | Ligand search using the PDB reference dictionary | http://www.ebi.ac.uk/msd-srv/chempdb |
| PDBeMotif | Query and analysis of structure, sequence motifs and interactions | http://www.ebi.ac.uk/pdbe-site/PDBeMotif/ |
| PDBePISA | Search and analysis of Protein Interfaces, Surfaces and Assemblies | http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html |
| PDBeFold | Secondary Structure Matching (SSM) service for comparing protein structures in 3D | http://www.ebi.ac.uk/msd-srv/ssm/ |
| PDBeTemplate | Search of local residue interactions in the PDB | http://www.ebi.ac.uk/pdbe-as/PDBeTemplate/ |
| PDBeAnalysis | Validation and analysis of PDBe data | http://www.ebi.ac.uk/pdbe-as/PDBeValidate |
| OLDERADO | Clustering information for NMR entries in the PDB | http://www.ebi.ac.uk/pdbe/olderado/ |
| PDBeMine | Supports ad-hoc queries and data analysis based on the relational PDBe database | http://www.ebi.ac.uk/pdbe-srv/msdmine |
These services can also be accessed through the main PDBe website, http://www.ebi.ac.uk/pdbe.
PDBeView ATLAS PAGES
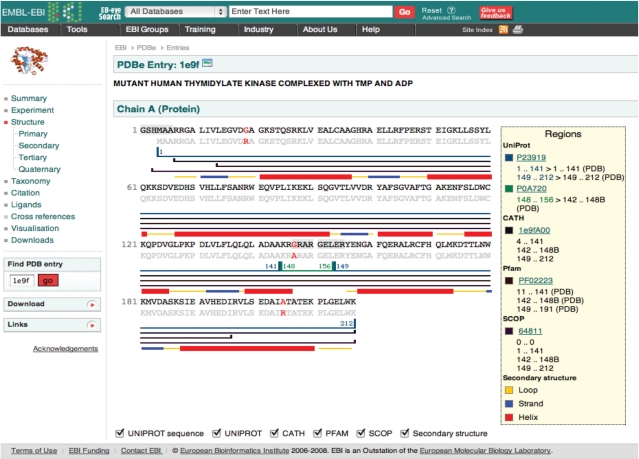
PDBeView Atlas pages (Figure 1) provide an overview of an individual PDB entry in a user-friendly layout and serve as a starting point for further browsing, searching or analysis. For each entry, the Atlas pages present all the information available in the database and provide:
a summary of the most important information for an entry together with high-quality images;
information about the primary, secondary, tertiary and quaternary structure of the macromolecules in the entry;
details about citations, taxonomy, experimental data, cross-references to other databases and ligand information;
access to interactive 3D visualization tools; and
access to file downloads.
Figure 1.
Example of an Atlas page, in this case for PDB entry 1E9F. The menus on the left-hand side enable navigation between different areas of information as well as links to other resources and downloadable files. The main panel on the right displays the sequence annotated with secondary structure information from various other databases (Uniprot, CATH, Pfam and SCOP).
Where available, the Atlas pages include hyperlinks directing the user to additional resources on the web such as UniProt, NCBI taxonomy and CiteXplore (http://www.ebi.ac.uk/citexplore/). The pages also provide a starting point for browsing the PDB with ‘magnifying glass’ links next to various terms and keywords. For example, by clicking on the magnifying glass icon next to an author name in the ‘Citation’ section, a list of all the PDB entries linked to that author will be obtained.
PDBeView SIMPLE SEARCH SYSTEM
PDBeView provides an easy-to-use search tool that serves as a starting point for simple textual searches based on Lucene search technology (http://lucene.apache.org/). The user can type any search term into the search box, e.g. author name, enzyme name or a ligand code. The hits are shown in tabular form and further filtering functionality is available, based on the provenance of the hits. For example, the search term ‘cat’ could be a 3-letter ligand code, an organism name or an author name. The result list shows the provenance of the hits found in various categories such as chemical compound, taxonomy, keywords or citation giving the user the option to refine the query.
PDBeView ADVANCED SEARCH SYSTEM
PDBeView also supports a more advanced form-based search interface that is suitable for more experienced PDB users and more complex queries. This interface enables users to search specifically on a number of database fields and to carry out sequence searches using FASTA (35). The search system also supports the sophisticated query expression language provided by the Lucene search engine. This allows users to specify fields and logical operators and build up complex queries (http://www.ebi.ac.uk/pdbe-srv/view/search_help). For instance, ‘method_class:x-ray and (chemical_code:atp or chemical_code:gtp) and (polymer_type:dna or polymer_type:rna)’ queries the PDB for entries that are determined using X-ray crystallography and contain at least one ligand molecule of type ATP or GTP in a complex involving at least one DNA or RNA molecule.
PDBeMotif
Database queries that include searches on 3D structural data are notoriously complex and computationally demanding. The complexity increases if the 3D queries are to be combined with sequence searches (similarity or patterns) or chemical fragment searches. To address these issues, the capabilities of our previous MSDsite (36), MSDmotif (37) and chemsearch (38) services have been combined in one powerful new service called PDBeMotif.
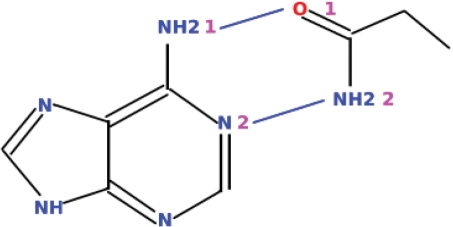
The core of this new system is a generic, metadata-driven query engine that forms the basis of multiple query interfaces, including the browser-based interface of PDBeMotif. A description of a user’s search is sent to a query engine in XML format. This engine then constructs an optimal SQL query on-the-fly and executes it on the specially designed PDBeMotif database. The results can be returned in a variety of manners, e.g. as an XML form or a series of HTML pages. The query engine can also act as a web service for Simple Object Access Protocol (SOAP)-based client applications. The software is available under General Public License at http://sourceforge.net/projects/pdbsam. Figure 2 shows a query example with which all instances in the PDB of a double hydrogen bond between an adenine fragment in a bound molecule and the side chain of an Asn or Gln residue in a protein could be obtained in seconds. The service provides examples (http://www.ebi.ac.uk/pdbe-site/pdbemotif/help/help.jsp?topic=eg) on how to define more complex queries such as building a ligand-specific sequence pattern based on the 3D interactions observed in the PDB or querying for protein domains that contain a binding site with protein–ligand interactions similar to those observed in a user-defined ligand binding site.
Figure 2.
Example of a graphically defined query that can be submitted to PDBeMotif.
BIOBAR
Biobar is a browser extension in the form of a toolbar for the open source web browser Firefox (http://www.mozilla.org). The latest version (2.0) has several improvements to functionality and usability including search capability for over 45 biological database collections, including NCBI, DDBJ, EBI, various structural databases (PDBe, RCSB, PDBsum, etc.), and other sequence, literature, plant, animal and function resources. The extensive main menu provides static links to more advanced bioinformatics analysis tools such as for structure and sequence alignment. The toolbar also provides hyperlinks for data-deposition sites for sequences and structure information. The search menu of Biobar is dynamic and changes based on the database that is to be queried. A detailed ‘options’ menu can be used to further customize the look-and-feel of the toolbar. Biobar 2.0 also offers a context-dependent literature search for any highlighted text on the browser page. Biobar is available for download from the Mozilla Add-ons pages (http://addons.mozilla.org/addon/169).
PDBeStatus
PDBeStatus shows the official status of any PDB entry that has been released or is undergoing processing at any of the wwPDB partner sites. The service allows users to subscribe to an RSS feed with the latest PDB release information or to define a search based on keywords. The results of the search are updated weekly and returned as a dynamic RSS feed.
PDBeFold
PDBeFold is based on the program SSM (39,40) and offers an interactive service for comparing protein structures in 3D. It is a powerful, flexible, fast and accurate tool for protein structure comparison that has rapidly gained popularity in the structural biology community. PDBeFold allows for 3D alignment of a query structure against individual protein chains, domains or the PDB and SCOP archives. The results are annotated with detailed secondary-structure alignments and the CATH and SCOP domain classifications. The results pages provide hyperlinks to related bioinformatics resources such as PDBeMotif, OCA, GeneCensus (41), FSSP (42), 3Dee (43), PDBsum (44), UniProt (9) and ProtoMap (45). The server provides interactive graphics capabilities to view aligned structures, using either RASMOL (http://rasmol.org/) or Jmol (http://jmol.sourceforge.net/). Studies have shown that the algorithm is particularly efficient (46), compared with similar resources, when applied to large protein structures (more than a few hundred amino acid residues) and when matching a structure to a database of structures (PDB, SCOP or user defined). The server also supports comparison and 3D alignment of multiple structures.
PDBePISA
PDBePISA (25) is an interactive service for prediction of probable quaternary states for crystal structures. The PDB archive stores the contents of the crystallographic asymmetric unit that may or may not describe the quaternary state of the molecules. Application of crystallographic symmetry and analysis of the interactions at the interfaces is necessary to detect all possible stable arrangements of the molecules in the crystal. It is important to understand the quaternary state (assemblies) when studying the biological function of macromolecules. The PDBePISA service provides information on all interfaces (protein, DNA/RNA and ligands) and allows searches for structurally similar interfaces and assemblies in the entire PDB. The assembly and interface information for the whole PDB archive is pre-compiled which makes these searches extremely fast. Users are also able to upload their own structural data in PDB or mmCIF format to obtain information on probable assemblies and structural and chemical properties of the interfaces. The results pages provide extensive analyses of the quaternary structure and residues involved in interactions at the interfaces.
PDBe DATABASE MAPPING (SIFTS)
The SIFTS initiative integrates data from a number of bioinformatics resources. A major obstacle to integration of PDB and UniProt, which are the primary archival databases for macromolecular structure and protein sequence data, respectively, is the absence of an up-to-date and well-maintained mapping between corresponding entries. PDBe works closely with the UniProt team at EBI to improve the taxonomy and sequence cross-reference information in the PDB and UniProt. This project began in 2001 and has resulted in a robust mechanism for exchanging data between the two resources. Based on the cross-referencing information between PDB and UniProt, information from databases such as Pfam, InterPro, CATH, SCOP, GO, IntEnz and PubMed is integrated and made available in XML format (ftp://ftp.ebi.ac.uk/pub/databases/msd/sifts/). The data are also exported in tab-delimited format with cross-references from PDB to SCOP, CATH, Pfam, PubMed, EC, NCBI taxonomy, UniProt, GO and InterPro. SIFTS data are released in synchronicity with the PDB release cycle. The present (September 2009) release of SIFTS contains 57 173 XML files compared with 59 330 entries in the PDB archive. Of the processed PDB entries, 55 872 entries have UniProt cross-reference information whereas sequence information from 3311 PDB entries is not available in UniProt. Figure 3 shows the process for generation of SIFTS XML files.
Figure 3.
Schematic overview of the process by which SIFTS files are generated (see text for details).
PDBeMapQuick
PDBeMapQuick is a service based on the data available in SIFTS. It provides a simple search mechanism based on PDB ID code and returns cross-reference information to other databases such as UniProt, CATH, SCOP, EC, GO, InterPro, NCBI-taxonomy and Pfam on the chain or residue level. The service also allows queries based on external database accession numbers or ID codes to obtain related structures in the PDB archive.
OUTREACH AND FEEDBACK
PDBe continues to present roadshows at various laboratories throughout Europe. These events provide us with an invaluable opportunity to present the current state of the PDBe databases and services and, more importantly, to obtain feedback and comments on the systems that we provide. These presentations and workshops will be continued, as part of our ongoing commitment to meet the needs and requirements of all our users in the bioscience communities.
PDBe AND NMR
Besides simplifying deposition, the main goals of the PDBe in relation to NMR are to provide software tools for NMR structures, and to make archived NMR data more accessible. As part of the latter effort, PDBe analyses archived NMR data to assess, improve or validate its quality and feed this data back to the community so that it can be used to improve existing or create new software.
PDBe continues its longstanding collaboration with BMRB on standardizing the NMR restraint data deposited at the PDB (47). This collection now contains over 5000 entries at BMRB that can be accessed directly for every NMR entry from the PDBeView Atlas pages under the ‘Experiment’ section. CCPN project files that contain both the restraints and the coordinates were generated as part of this restraint-standardization process. Links are available to both the original converted restraints (DOCR) and the filtered restraints (FRED), where redundant information has been removed.
Another focus of the NMR effort at PDBe is large-scale data analysis. A framework for processing archived data now exists that is centered around information in CCPN projects. This framework was originally developed as part of the restraint-standardization process and resulted in a study where the overall information content of NMR distance restraints was compared with the deposited coordinates (48). The results are available on-line (http://www.ebi.ac.uk/pdbe/docs/NMR/analysis/results/html/). The processing framework was further developed to link chemical shift information to the atomic coordinate data in the PDB. Data from 1959 BMRB entries was analysed and revealed useful relationships between the per-atom accessible surface area (as calculated from the coordinates) and the chemical shift dispersion (49). Such information is potentially very useful for chemical-shift validation and it is available on-line (http://www.ebi.ac.uk/pdbe/docs/NMR/shiftAnalysis/).
In collaboration with CCPN and SpronkNMR consultancy, PDBe has developed an on-line version of the FormatConverter software (19). This service is available from http://webapps.ccpn.ac.uk/fcweb/ and enables users to upload NMR data and coordinate files to create a CCPN project. Formats supported include ANSIG, ARIA, AutoAssign, CNS/XPLOR, CYANA, FASTA, MARS, MODULE, MOLMOL, MONTE, NmrDraw, NmrView, PDB, Pronto, SHIFTX, Sparky, TALOS and XEASY.
The remediated wwPDB library of ligands, small molecules and monomers (22) have been converted to the new CCPN version 2.0 framework and is available as CCPN XML files (http://www.ebi.ac.uk/pdbe/docs/NMR/chemCompXml_2.0/main.html). The information was also converted to reference files available in tabulated and XML formats (http://www.ebi.ac.uk/pdbe/docs/NMR/refData_2.0/main.html). These files are free to download and use, although we request that any changes or mistakes are reported so they can be fed back to the community.
PDBe has been involved in the development of a new method called Complementary Coordinates (COCO) (50) for the analysis of ensembles of NMR structures. This method analyses the distribution of an NMR ensemble in conformational space, and generates new models for this ensemble that fill ‘gaps’ in the existing distribution. COCO should therefore be a useful aid in NMR structure refinement and in other situations where a richer representation of conformational variability is desired, for example in docking studies. COCO is freely accessible via the website http://www.ccpb.ac.uk/software/COCO.
PDBe provides access to the OLDERADO (51) database, which contains a list of the most representative models in an NMR ensemble after clustering. Since the summer of 2008, all new NMR entries in the PDB are processed with the original OLDERADO protocol. This uses NMRCORE to define the core atoms in the ensemble and the rigid domains within the protein (52), and NMRCLUST to identify conformationally related subfamilies from this information (53). Due to restrictions in this protocol, it handles only protein NMR structures containing a well-defined core, and the list of representative models is currently not available for DNA or RNA structures, short flexible proteins or ligand complexes. The data are accessible from the PDBeView Atlas pages or can be accessed directly from the OLDERADO site at PDBe (http://www.ebi.ac.uk/pdbe/olderado/).
THE ELECTRON MICROSCOPY DATA BANK
EMDB is the international repository for density maps derived by high-resolution biological transmission EM. EMDB was originally established at the EBI in 2002 and is now developed and operated jointly with RCSB and Baylor College of Medicine with NIH funding. EMDB contains both macromolecular structures reconstructed using the single-particle method and images of sub-cellular regions from electron tomography. At present (September 2009), EMDB contains 701 entries. EMDB operates a web-based deposition (26) and retrieval system with manual curation. Maps are submitted through the web interface and reviewed by the annotation staff at PDBe or RCSB, before they are made available in a weekly release cycle at both sites through a search system and FTP access.
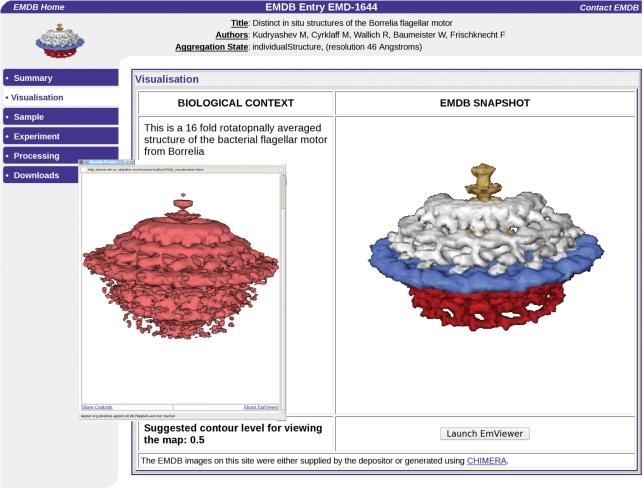
An important goal of this project is to bring about a close integration of maps in EMDB and the corresponding 3D structures in the PDB. This has been partly achieved by providing a direct path from an EMDB map deposition to a PDB deposition of 3D structures fitted into the map. The visualization of EMDB maps has been improved by adding Java applets to the EMDB Atlas pages that allow for interactive generation of isosurface representations (Figure 4). This can be done both with the OpenAstexViewer (http://openastexviewer.net/web/), an open source version of the AstexViewer package that has been used at PDBe for a long time and with a new applet called EMviewer developed at Baylor College of Medicine and Rice University.
Figure 4.
The new EMViewer 3D visualization Java applet is available on the EMDB Atlas pages and allows interactive generation of isosurface representations. The map shown here is of the Borrelia flagellar motor, accession number EMDB-1644 (Authors: Kudryashev, M., Cyrklaff, M., Wallich, R., Baumeister, W., Frischknecht, F.).
To make EMDB more easily accessible to users, we have started the development of a new website, http://www.emdatabank.org/, hosted jointly by PDBe and RCSB. This site will serve as the central portal for deposition and retrieval services. A first example of new services is the ‘recent releases’ page that gives a weekly overview of new maps available in the EMDB database.
FUTURE DEVELOPMENTS
PDBe continues to work closely with its collaborators (wwPDB partners, structural biology community, bioinformatics resources at EBI and elsewhere) to improve the quality and consistency of the data in the PDB. Considerable efforts are also put into improving PDBe services and tools, and we are currently working towards a streamlined, unified interface to these services. The search, browse and delivery capabilities at PDBe are under active development, and many new features, services and tools can be expected in the next couple of years. The underlying relational database continues to be improved, with new features and capabilities being added to many services, moving us ever closer to our ultimate goal of becoming a comprehensive, integrated resource for structural information on bio-macromolecules.
FUNDING
Wellcome Trust (GR062025MA and 088944); European Union (TEMBLOR, FP6 Extend-NMR and 3D-EM NoE); CCP4; Biotechnology and Biological Sciences Research Council; Medical Research Council; NIH; European Molecular Biology Laboratory. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to thank all collaborators and partners in the EBI, EMBL, wwPDB and other collaborative efforts, as well as the structural biology community for depositing their experimental data in the PDB and EMDB.
REFERENCES
- 1.Tagari M, Tate J, Swaminathan GJ, Newman R, Naim A, Vranken W, Kapopoulou A, Hussain A, Fillon J, Henrick K, et al. E-MSD: improving data deposition and structure quality. Nucleic Acids Res. 2006;34:D287–D290. doi: 10.1093/nar/gkj163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kouranov A, Xie L, de la Cruz J, Chen L, Westbrook J, Bourne PE, Berman HM. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 6.Berman HM. The Protein Data Bank: a historical perspective. Acta Cryst. 2008;A64:88–95. doi: 10.1107/S0108767307035623. [DOI] [PubMed] [Google Scholar]
- 7.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Consortium,UniProt. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das D, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrell D, Dimmer E, Huntley R, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009—an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebbert JO, Dupras DM, Erwin PJ. Searching the medical literature using PubMed: a tutorial. Mayo Clin. Proc. 2003;78:87–91. doi: 10.4065/78.1.87. [DOI] [PubMed] [Google Scholar]
- 15.Velankar S, McNeil P, Mittard-Runte V, Suarez A, Barrell D, Apweiler R, Henrick K. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2005;33:D262–D265. doi: 10.1093/nar/gki058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogh R, Ionides J, Ulrich E, Boucher W, Vranken WF, Linge JP, Habeck M, Rieping W, Bhat TN, Westbrook J, et al. The CCPN project: an interim report on a data model for the NMR community. Nat. Struct. Biol. 2002;9:416–418. doi: 10.1038/nsb0602-416. [DOI] [PubMed] [Google Scholar]
- 17.Fogh R, Boucher W, Vranken WF, Pajon A, Stevens T, Bhat TN, Westbrook J, Ionides J, Laue ED. A framework for scientific data modeling and automated software development. Bioinformatics. 2005;21:1678–1684. doi: 10.1093/bioinformatics/bti234. [DOI] [PubMed] [Google Scholar]
- 18.Fogh R, Vranken WF, Boucher W, Stevens T, Laue ED. A nomenclature and data model to describe NMR experiments. J. Biomol. NMR. 2006;36:147–155. doi: 10.1007/s10858-006-9076-z. [DOI] [PubMed] [Google Scholar]
- 19.Vranken WF, Boucher W, Stevens T, Fogh R, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 20.Tagari M, Newman R, Chagoyen M, Carazo J, Henrick K. New electron microscopy database and deposition system. Trends Biochem. Sci. 2002;27:589. doi: 10.1016/s0968-0004(02)02176-x. [DOI] [PubMed] [Google Scholar]
- 21.Swedlow JR, Goldberg IG, Eliceiri KW the OME Consortium. Bioimage informatics for experimental biology. Annu. Rev. Biophys. 2009;38:327–346. doi: 10.1146/annurev.biophys.050708.133641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henrick K, Feng Z, Bluhm WF, Dimitropoulos D, Doreleijers JF, Dutta S, Flippen-Anderson JL, Ionides J, Kamada C, Krissinel E, et al. Remediation of the protein data bank archive. Nucleic Acids Res. 2008;36:D426–D433. doi: 10.1093/nar/gkm937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tagari M, Tate J, Swaminathan GJ, Newman R, Naim A, Vranken WF, Kapopoulou A, Hussain A, Fillon J, Henrick K, et al. E-MSD: improving data deposition and structure quality. Nucleic Acids Res. 2006;34:D287–D290. doi: 10.1093/nar/gkj163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleywegt GJ, Harris MR, Zou JY, Taylor TC, Wählby A, Jones TA. ‘The Uppsala Electron-Density Server’. Acta Cryst. 2004;D60:2240–2249. doi: 10.1107/S0907444904013253. [DOI] [PubMed] [Google Scholar]
- 25.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Henrick K, Newman R, Tagari M, Chagoyen M. EMDep: a web-based system for the deposition and validation of high-resolution electron microscopy macromolecular structural information. J. Struct. Biol. 2003;144:228–237. doi: 10.1016/j.jsb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milner-White EJ, Russell MJ. Sites for phosphates and iron-sulfur thiolates in the first membranes:3 to 6 residue anion-binding motifs (nests) Orig Life Evol. Biosph. 2005;35:19–27. doi: 10.1007/s11084-005-4582-7. [DOI] [PubMed] [Google Scholar]
- 29.Watson JD, Milner-White EJ. A novel main-chain anion-binding site in proteins: the nest a particular combination of φ/ψ values in successive residue gives rise to anion-binding sites that occur commonly and are found often at functionally important regions. J. Mol. Biol. 2001;315:171–182. doi: 10.1006/jmbi.2001.5227. [DOI] [PubMed] [Google Scholar]
- 30.Milner-White EJ. Beta-bulges within loops as recurring features of protein structure. Biochimica et Biophysica Acta. 1987;911:261–265. doi: 10.1016/0167-4838(87)90017-3. [DOI] [PubMed] [Google Scholar]
- 31.Questel JYL, Morris DG, Maccallum PH, Poet R, Milner-White EJ. Common ring motifs in proteins involving asparagine or glutamine amide groups hydrogen-bonded to main-chain atoms. J. Mol. Biol. 1993;231:888–896. doi: 10.1006/jmbi.1993.1335. [DOI] [PubMed] [Google Scholar]
- 32.Watson JD, Milner-White EJ. The conformations of polypeptide chains where the main-chain parts of successive residues are enantiomeric. Their occurrence in cation and anion-binding regions of proteins. J. Mol. Biol. 2002;315:183–191. doi: 10.1006/jmbi.2001.5228. [DOI] [PubMed] [Google Scholar]
- 33.Milner-White EJ, Poet R. Loops, bulges, turns and hairpins in proteins. Trends Biochem. Sci. 1987;12:189–192. [Google Scholar]
- 34.Duddy WJ, Nissink JWM, Allen FH, Milner-White EJ. Mimicry by asx- and ST-turns of the four main types of beta-turn in proteins. Protein Sci. 2004;13:3051–3055. doi: 10.1110/ps.04920904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 36.Golovin A, Dimitropoulos D, Oldfield T, Rachedi A, Henrick K. MSDsite: a database search and retrieval system for the analysis and viewing of bound ligands and active sites. PROT. Struct. Funct. Bioinformatics. 2005;58:190–199. doi: 10.1002/prot.20288. [DOI] [PubMed] [Google Scholar]
- 37.Golovin A, Henrick K. MSDmotif: exploring protein sites and motifs. BMC Bioinformatics. 2008;9:312. doi: 10.1186/1471-2105-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golovin A, Henrick K. Chemical substructure search in SQL, J. Chem. Inf. Model. 2009;49:22–27. doi: 10.1021/ci8003013. [DOI] [PubMed] [Google Scholar]
- 39.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Cryst. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. Common subgraph isomorphism detection by backtracking search. Softw. Pract. Exp. 2004;34:591–607. [Google Scholar]
- 41.Lin J, Qian J, Greenbaum D, Bertone P, Das R, Echols N, Senes A, Stenger B, Gerstein M. GeneCensus: genome comparisons in terms of metabolic pathway activity and protein family sharing. Nucleic Acids Res. 2002;30:4574–4582. doi: 10.1093/nar/gkf555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siddiqui AS, Dengler U, Barton GJ. 3Dee: a database of protein structural domains. Bioinformatics. 2001;17:200–201. doi: 10.1093/bioinformatics/17.2.200. [DOI] [PubMed] [Google Scholar]
- 44.Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yona G, Linial N, Linial M. ProtoMap: automatic classification of protein sequences and hierarchy of protein families. Nucleic Acids Res. 2000;28:49–55. doi: 10.1093/nar/28.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolodny R, Koehl P, Levitt M. Comprehensive evaluation of protein structure alignment methods: scoring by geometric measures. J. Mol. Biol. 2005;346:1173–1188. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doreleijers JF, Nederveen AJ, Vranken WF, Lin J, Bonvin AMJJ, Kaptein R, Markley JL, Ulrich EL. BioMagResBank databases DOCR and FRED containing converted and filtered sets of experimental NMR restraints and coordinates from over 500 protein PDB structures. J. Biomol. NMR. 2005;32:1–12. doi: 10.1007/s10858-005-2195-0. [DOI] [PubMed] [Google Scholar]
- 48.Vranken WF. A global analysis of NMR distance constraints from the PDB. J. Biomol. NMR. 2007;39:303–314. doi: 10.1007/s10858-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vranken WF, Rieping W. Relationship between chemical shift value and accessible surface area for all amino acid atoms. BMC Struct.Biol. 2009;9:20. doi: 10.1186/1472-6807-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laughton CA, Orozco M, Vranken WF. COCO: a simple tool to enrich the representation of conformational variability in NMR structures. Proteins. 2008;75:206–216. doi: 10.1002/prot.22235. [DOI] [PubMed] [Google Scholar]
- 51.Kelley LA, Sutcliffe MJ. OLDERADO: on-line database of ensemble representatives and domains. On Line Database of Ensemble Representatives And DOmains. Protein Sci. 1997;6:2628–2630. doi: 10.1002/pro.5560061215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelley LA, Gardner SP, Sutcliffe MJ. An automated approach for defining core atoms and domains in an ensemble of NMR-derived protein structures. Protein Eng. 1997;10:737–741. doi: 10.1093/protein/10.6.737. [DOI] [PubMed] [Google Scholar]
- 53.Kelley LA, Gardner P, Sutcliffe MJ. An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally-related subfamilies. Protein Eng. 1996;9:1063–1065. doi: 10.1093/protein/9.11.1063. [DOI] [PubMed] [Google Scholar]