Abstract
3DNALandscapes, located at: http://3DNAscapes.rutgers.edu, is a new database for exploring the conformational features of DNA. In contrast to most structural databases, which archive the Cartesian coordinates and/or derived parameters and images for individual structures, 3DNALandscapes enables searches of conformational information across multiple structures. The database contains a wide variety of structural parameters and molecular images, computed with the 3DNA software package and known to be useful for characterizing and understanding the sequence-dependent spatial arrangements of the DNA sugar-phosphate backbone, sugar-base side groups, base pairs, base-pair steps, groove structure, etc. The data comprise all DNA-containing structures—both free and bound to proteins, drugs and other ligands—currently available in the Protein Data Bank. The web interface allows the user to link, report, plot and analyze this information from numerous perspectives and thereby gain insight into DNA conformation, deformability and interactions in different sequence and structural contexts. The data accumulated from known, well-resolved DNA structures can serve as useful benchmarks for the analysis and simulation of new structures. The collective data can also help to understand how DNA deforms in response to proteins and other molecules and undergoes conformational rearrangements.
INTRODUCTION
In addition to the genetic message, DNA base sequence carries a multitude of structural and energetic signals related to its biological packaging and processing. These codes govern how the double-helical molecule deforms in response to proteins and other ligands and when and where the genetic information is expressed. DNA is not just a passive substrate of cellular proteins but an active player with physical properties capable of influencing the three-dimensional organization of genetic sequences and the activity of regulatory proteins and processing enzymes. Understanding the pathways and capabilities of DNA deformation is thus crucial for deciphering the codes behind the regulation, organization and dynamics of various genomes. Acquiring this knowledge requires a systematic view of the structural landscapes accessible to DNA as it deforms in solution and adjusts to interactions with other molecules. This information, in turn, offers reliable benchmarks for predictions of nucleic-acid interactions and structures.
3DNALandscapes is a new database for exploring the conformational features of DNA. The database has been designed to study DNA backbone, side-group, base-pair, base-pair-step and complementary-strand geometry statistically, using information derived from multiple structures with the 3DNA software package (1–3) in combination with other currently available data resources, such as structural classifications and descriptions found in the Protein Data Bank (PDB) (4) and Nucleic Acid Database (NDB) (5). We have also constructed a web interface to link, report, plot and analyze the structural parameters in the database. The main component of the web interface is a search function that enables the user to collect structural data and generate statistical reports on the fly.
The PDB and NDB contain a number of derived nucleic-acid conformational parameters, including the base-pair and base-pair-step parameters obtained with 3DNA. Although these databases include some of the information stored in 3DNALandscapes, not all of the information is contained in either of them. In addition, the PDB and NDB are designed to be structure-centric, meaning that data from a single structure are easy to obtain. Gathering data for a specific parameter or parameter set across multiple nucleic-acid structures is difficult or impossible with these interfaces. The collective information in 3DNALandscapes provides insights into the intrinsic sequence-dependent structure and deformability of DNA (6,7) as well as useful benchmarks for the analysis and simulation of other DNA structures (8–10).
DATABASE CONTENT
The database is managed by a MySQL platform (11). Data are stored in a rational schema that organizes tables of information in a hierarchical fashion. The highest level of the schema contains basic structural information, such as molecular classifications, sequences and resolution. The next level divides the data into five categories: backbone, sugar-base side-group, base-pairing, base-pair-step and complementary-strand information. The lowest level of the schema contains the derived parameters associated with the backbones, side groups, base pairs, base-pair steps and complementary-strand interactions.
Structures
The first release of the database contains derived information for all DNA-containing structures—both free and bound to proteins, drugs and other ligands—deposited in the Protein Data Bank as of October 2009. The composite data come from 6615 structural models, taken from the complete sets of atomic coordinates reported in 2084 X-ray crystallographic and 586 nuclear magnetic resonance (NMR) investigations. Among those structures, 1429 occur in complexes with proteins, 973 associate with drugs and other small molecules and 2004 contain only bound water or metal ions. The X-ray-based entries reflect the coordinates of the biological units rather than the asymmetric structural units. Individual models within the ensembles of NMR-derived structures contain unique internal identifiers assigned as the database is loaded.
The structures are classified in terms of the DNA conformational assignments made by the 3DNA software, e.g. fraction or number of base-pair steps in A and B double-helical forms. Individual entries also include the resolution (in the case of X-ray models), literature citations and other features stored in the original structural files. The DNA sequences and associated chain names and residue numbers are extracted in the 3DNA analysis for subsequent use in locating specific nucleotides, base pairs and base-pair steps in a given model. The data-collection procedure records the chemical composition and nucleotide surroundings of the base pairs and base-pair steps so that effects of base modification and sequence context can be studied. That is, the base-pair and dimeric entries contain the identities of the base pairs that precede and follow the designated unit, thereby marking the relevant set of conformational data in the context of the trimer that contains the base pair and the tetramer than contains the dimer step. The annotation thus takes account of the base pairs and base-pair steps at the ends of helices.
Backbones
Features of the DNA chemical framework stored in the database include the standard set of internal torsional parameters associated with the nucleotide units along individual strands (12) and related intrastrand distances. These quantities include the five acyclic torsion angles—α (O3′–P–O5′–C5′), β (P–O5′–C5′–C4′), γ (O5′–C5′–C4′–C3′), δ (C5′–C4′–C3′–O3′), ε (C4′–C3′–O3′–P), ζ (C3′–O3′–P–O5′)— along the sugar-phosphate backbone and the distances dP–Pbetween phosphorus atoms on successive nucleotides. The distances are expressed in angstrom units and the angles are assigned values over the range (−180°, +180°).
Sugar-base side groups
Description of the spatial arrangements of the sugar and base units follows conventional guidelines (12). The stored conformational data include: (i) the glycosyl torsion angle χ (O4′–C1′–N9–C4) or χ (O4′–C1′–N1–C2), respectively, describing the orientation of a purine (R) or pyrimidine (Y) with respect to the sugar ring; (ii) the five internal sugar-ring torsion angles—ν0 (C4′–O4′–C1′–C2′), ν1 (O4′–C1′–C2′–C3′), ν2 (C1′–C2′–C3′–C4′), ν3 (C2′–C3′–C4′–O4′) and ν4 (C3′–C4′–O4′–C1′); and (iii) the phase angle P and amplitude τmax of sugar pseudorotation derived from the latter quantities (13).
Base pairs
The 3DNA analysis identifies 91 280 hydrogen-bonded base pairs—70 120 canonical (Watson–Crick) pairs and 21 160 noncanonical pairs—in the above set of structures. The Watson–Crick pairs include all A·T and G·C associations with the requisite hydrogen-bond (H-bond) patterns. All other base pairs, including partially distorted Watson–Crick pairs with missing H bonds, are classified as noncanonical. Structures with three or more strands include the close base-base associations of all interacting strands. The accepted base pairs meet simple geometric criteria (14) and contain two or more H bonds, at least one of which involves a proton donor–acceptor interaction between nitrogens or oxygens on the two bases.
The spatial disposition of the bases in each pair is described by three types of data: (i) the identities and lengths of the H bonds; (ii) the six rigid-body parameters that relate local coordinate frames embedded on the interacting bases; and (iii) the virtual distances and angles between selected atoms on the bases and attached sugars. The set of H bonds includes the interactions between the flagged bases as well as those with the sugar-phosphate backbone and the bifurcated (three-center) H-bonds between contacted residues. The base-pair parameters—three angles called Buckle, Propeller and Opening and three distances called Shear, Stretch and Stagger (15)—follow the matrix-based definitions originated by Zhurkin et al. (16) and described in detail by El Hassan and Calladine (17). The virtual parameters include the distances dC1′···C1′ between the C1′ atoms attached to paired bases and the angles λR and λY formed by the C1′···C1′ line with the R(C1′–N9) and Y(C1′–N1) glycosidic bonds, respectively.
Base-pair steps
Structural characterization of the 66 549 base-pair steps formed by sequential base pairs includes: (i) the six rigid-body parameters specifying the orientation and displacement of the constituent base pairs; (ii) the six local helical parameters relating the positions of the base pairs; (iii) the area of overlap of the stacked base pairs; (iv) the displacement of the phosphorus atoms on interacting strands along the local dimeric and helical coordinate frames; (v) the distances between the C1′ atoms in the dimeric unit; and (vi) the conformational families to which the steps belongs. The coordinate frames on the bases, base pairs and base-pair steps follow established conventions (18). The six base-pair-step parameters—three rotations (Tilt, Roll, Twist) and three translations (Shift, Slide, Rise) (15)—are analogs of the six base-pair parameters (16,17). The six local helical parameters—Inclination, Tip, Helical Twist, x-displacement, y-displacement and Helical Rise (15)—are defined, following Babcock et al. (19), in terms of the single rotational operation that brings the coordinate frames on the base pairs into alignment. The base-pair overlap is the area shared by the four polygons formed by projecting the ring atoms of the bases on the mean base-pair plane (1). The stored data include the contributions to the overlap from the bases on the same and opposing strands and the corresponding values obtained for larger polygons constructed from the ring and exocyclic base atoms. The projections of the P atoms (xP, yP, zP) along the coordinate axes of the dimeric step distinguish A- from B-type DNA (7) as well as potential intermediate AB steps along the A→B conformational pathway (10). The corresponding projections along the axes of the local helical frame [xP(h), yP(h), zP(h)] distinguish the TA-like steps (1), i.e. the conformational form of DNA (20) found in complexes with the TATA-box protein and other proteins. The zP and zP(h) values are used to determine the conformational family of the dimer steps. The intrastrand C1′···C1′ distances also distinguish different conformational types.
Complementary-strand interactions
Finally, the conformational data include the widths of the major and minor grooves, i.e. the long-range distances between phosphorus atoms on interacting strands that expose the respective non-H-bonded edges of Watson–Crick base pairs. The recorded values are based on the direct and refined formulations of El Hassan and Calladine (21) and are assigned to the relevant base-pair step. The direct values correspond to the distances between Pi, the phosphorus atom on the leading strand of base-pair step i, and specific phosphorus atoms on the other strand, Pi−3 across the minor groove and Pi+4 across the major groove, typically the shortest cross-strand P···P distances in B-DNA helices. The refined values allow for the variation in helical structure that alters the identities of the atoms in closest cross-strand contact. Thus, the two measures of groove width may differ markedly if the helix undergoes large distortions.
WEB INTERFACE
The web interface, located at: http://3DNAscapes.rutgers.edu and constructed in the CodeIgniter PHP web application framework (22), parallels the organization of the database. The software contains three major components: a structure filter; a series of parameter- and context-selection panels; and a data report. The tabulated data also include links to a local summary and visualization page for each of the structural fragments from which the listed quantities are extracted. The user must first specify a set of structures in the structure filter, then select the type of structural information to be considered and finally view the summaries of the analysis in the statistical reporter.
Structure filter
The structure-filter page offers two options for the user to define a set of structures. First, one can make selections based on a combination of the following features: the experimental method used to determine the structure; the molecular contents; the resolution cutoff; and the conformational characteristics of the constituent base-pair steps. By specifying the experimental method, the user can examine structures obtained by X-ray, NMR or both approaches. The choice of molecular contents refers to the other molecules present in the experimental structure: proteins; drugs or other small molecules; bound water; metal ions. The conformational option allows the user to select structures with given fractions or numbers of base-pair steps that have local conformational features characteristic of A-, B-, AB-, TA- or Z-type helices. That is, the structure-filtering algorithm uses the values of various parameters, determined with the 3DNA software, to characterize individual base-pair steps in a given structure rather than group the structure as a whole in terms of its global appearance. Thus, A-type base-pair steps might occur in what appears at the global level to be a B-DNA duplex and vice versa. This information is useful in understanding how ligands induce local conformational changes in DNA or how large-scale reorganization of structure preserves fundamental local structural propensities. The resolution cutoff affects only the collection of X-ray structures. The NMR structures in the database have an arbitrarily assigned resolution of zero, which will lie always within the cutoff limit.
The second option lets the user enter a list of PDB or NDB structural identifiers (IDs), which the server checks for accuracy. This option allows the user to perform searches elsewhere, such as the integrated search at the NDB or the advanced search at the PDB, and then import the findings into the 3DNALandscapes interface for conformational analysis. After clicking ‘next’, a list of structures with brief descriptions is displayed in a table with sorting and paging capabilities. The user can edit the structures generated in the automated search by denoting the PDB identifiers of the files to be removed or added.
Finally, the user has the option in either selection process of choosing a representative structure (the first structure) or the complete ensemble of structures associated with the NMR-based files. It worth noting that the selection of ensembles can lead to time delays in the analysis and visualization of large quantities of data and also may bias the statistical results. The choice can be useful, however, if the user is interested in the conformational trends associated with the DNA included in a single NMR structure file.
Parameter- and context-selection panels
The parameter- and context-selection panels allow the user to choose the conformational parameters of interest and the nucleotide units that meet certain conditions within the set of selected structures. The parameter list includes the aforementioned quantities associated with the DNA backbones, sugar-base side groups, base pairs, base-pair steps and complementary strands. The set of conditions includes the chemical context, sequence context and conformational category.
Thus, the user can specify whether not to include nucleotides containing modified bases or those found in non-canonical base pairs. One can also select the identities of the bases that flank particular chemical moieties, such as the base pairs that precede and follow a base pair or base-pair step. Only parameters associated with the specified chemical unit in the given sequential context are retained. This option allows the user to study the effects of neighboring base pairs on the local conformation of DNA. The user can also narrow the search by specifying the conformational character of base-pair steps. This action restricts the selection of parameters to the backbones and base pairs that constitute the base-pair steps of a particular conformational type, e.g. only A-DNA steps (as opposed to the structures with a given proportion or number of A-like steps, which can be chosen with the Structure Filter).
Data report
The data report contains a table of the selected conformational data, a gallery of plotted images and a brief statistical report. The grid-view table at the top of the report lists all entries for the chosen parameters and contains hyperlinks, which direct the user to the local summary and visualization pages described below. The information in the table can be sorted by column entries and exported as a data file. The graphical gallery includes histograms and, in some cases, scatter plots of the distribution of the collected data. The histograms (Figure 1) illustrate the information included in individual columns, while the scatter plots (Figure 2) reveal the pairwise correlations of selected parameters, such as the coupling of bending and twisting in DNA base-pair steps (via Roll and Twist) (9). The scatter plots also include ellipses, derived from the covariance, that encircle most of the plotted data (6). Related parameters are plotted on a common scale for ease of comparison, and all images can be downloaded. The statistical report includes the number of examples, average values, minima and maxima for the data associated with the chosen sequences, such as the rigid-body parameters of specific base pairs or base-pair steps (Figure 1). The report also includes the option to determine the statistics for the chosen parameters in different trimeric or tetrameric sequence contexts. The analyses of rigid-body parameters of both base pairs and base-pair steps include the covariance matrices and derived sequence-dependent elastic constants. These knowledge-based parameters can be used to study many DNA bending and packaging problems, such as DNA cyclization (8) and nucleosome-positioning (9,10) propensities.
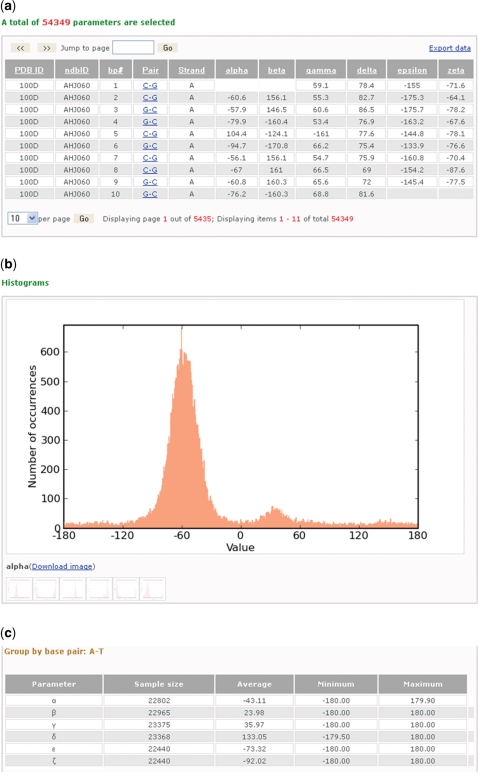
Figure 1.
Screenshots illustrating some of the information about the DNA sugar-phosphate torsion angles collected from a search of the paired nucleotides in all DNA-containing crystal structures of 3 Å or better resolution. (a) A table of 54 349 entries from 1895 different structures arranged in seven columns that respectively list the Protein Data Bank and Nucleic Acid Database identifiers of the structures (PDB ID, NDB ID), the residue number (bp#), the chemical identities of the paired bases (Pair), the strand identity of the first of the two listed bases (Strand) and the values of the six torsion angles (α, β, γ, δ, ξ and ζ) in the specified nucleotides. The data can be sorted by clicking on the headers of the columns and also exported into a tab-delimited file. (b) A close-up of one of the downloadable histograms—here the distribution of the torsion angle α about the O3′–P–O5′–C5′ chemical bond sequence—automatically generated for each of the angles in the above data set. Moving the mouse across the different icons reveals the corresponding distributions for the other angles. (c) Summary of statistical information, including the number of examples, average values, minima and maxima for the torsion angles in the data set. The report is divided into groups based on the type and composition of base pairs, here the canonical A·T Watson–Crick pair.
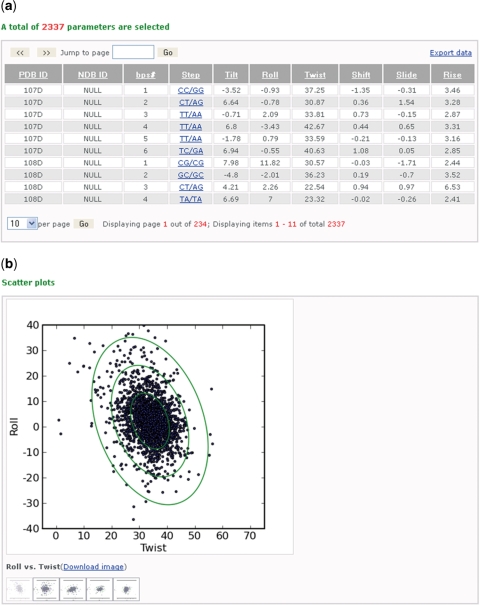
Figure 2.
Screenshots showing some of the information about the arrangements of DNA base-pair steps extracted from a search of all DNA-containing structures derived by solution NMR spectroscopic measurements. (a) A table of 2337 sets of base-pair-step parameters relating unmodified base pairs in 586 representative models from 586 PDB files arranged in columns that respectively list the PDB identifiers (PDB ID), the base-pair-step numbers (bps#), the chemical identities (Step) and the (Tilt, Roll, Twist, Shift, Slide and Rise) values describing the steps. Data can be manipulated and downloaded as described in Figure 1. (b) One of the downloadable two-dimensional scatter plots, here Roll versus Twist, automatically generated for the above steps in the data set. Ellipses are projections of six-dimensional ‘equipotential’ surfaces derived, following (6), from the plotted data. Contours correspond to ‘energies’ where parameters deviate from mean values by no more than n times the root-mean-square deviation, where n = 1–3. Moving the mouse across the different icons reveals four other such plots: Roll versus Slide; Twist versus Slide; Tilt versus Shift; Twist versus Rise.
Local summary and visualization
The local summary and visualization pages (Figure 3) give a detailed listing of the sequence context, H-bonding interactions, conformational parameters and atomic-level representations of each of the base pairs or base-pair steps incorporated in the data report. Each page contains three sections. The first section gives the complete sequence, the location(s) of the selected base pair(s), the number of H bonds between paired bases and the base-pair types (Watson–Crick or noncanonical) in the structural example. The second section lists the values of all conformational parameters associated with the given base pair or base-pair step, including the torsional angles about the glycosidic linkage and the attached sugar-phosphate backbones. The last section contains a two-dimensional stacking diagram of the base pair or base-pair step generated with 3DNA and a link to three-dimensional visualization and manipulation of the same unit with the JAVA-based Jmol software (23).
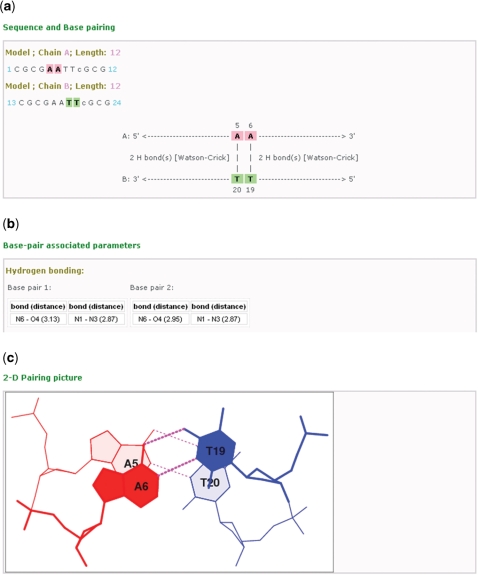
Figure 3.
Screenshots showing some of the data provided in the local summary and visualization pages of each of the base-pair steps, here the A5A6 ·T19T20 dimer from the 2.25-Å crystal structure of the Dickerson–Drew dodecamer complexed to netropsin (PDB ID: 101D) (25). (a) A map of the interactions and locations of the paired bases in the selected base-pair step, including information on the number of hydrogen bonds and type of base pair. (b) A summary of some of conformational parameters associated with the base-pair step, here the identities and lengths of the hydrogen bonds in each base pair. (c) A downloadable stacking diagram that illustrates the overlap and hydrogen-bonding patterns of the base pairs (A5·T20 and A6·T19) in the selected step.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
3DNALandscapes allows a user to gather information and gain insight about DNA sequence-dependent conformation and deformability from known-high-resolution structures. In contrast to other structural databases (4,5), which archive the Cartesian coordinates and/or derived parameters for individual structures, 3DNALandscapes enables searches and summarizes conformational data from multiple structures that meet selected criteria. To the best of our knowledge, there are no other databases with these unique capabilities.
The information collected in 3DNALandscapes also provides useful benchmarks for the analysis and simulation of other DNA structures. The data that characterize existing structures can be compared with new experimentally derived or computer-simulated DNA structures. The database can be used in combination with the 3DNA software tools (1,2) or the w3DNA web interface for such analyses (3). The knowledge-based potentials provided through 3DNALandscapes can be used in various computer applications, such as the simulation of fluctuating DNA polymers (8,24) or the analysis of nucleosome positioning on DNA (9,10). The access to large volumes of derived conformational information may stimulate new types of analyses and lead to new understanding of DNA structure and deformability.
We plan to connect 3DNALandscapes to the w3DNA server. We are currently investigating ways to identify nonredundant DNA-containing structures automatically and will include this information in future releases of 3DNALandscapes. We will update the database at regular intervals as new structures are added to the Protein Data Bank and Nucleic Acid Database.
FUNDING
The U.S. Public Health Service (research grants GM20861 and GM34809, instrumentation grant RR022375 and New Interdisciplinary Research Workforce Training Grant DK071502 (to G.Z.)). Partial funding for open access: U.S.P.H.S. grant GM3409 and the Mary I. Bunting Fund of Rutgers, the State University of New Jersey.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to thank Thomas Gaillard and Difei Wang for testing the database web interface and offering valuable comments. We also thank Thomas Chapin for lasting help in setting up the web server and database configuration.
REFERENCES
- 1.Lu X-J, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu X-J, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three dimensional nucleic-acid structures. Nat. Protoc. 2008;37:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng G, Lu X-J, Olson WK. Web 3DNA—a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009;37:W240–W246. doi: 10.1093/nar/gkp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh S-H, Srinivasan AR, Schneider B. The Nucleic Acid Database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olson WK, Gorin AA, Lu X-J, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu X-J, Shakked Z, Olson WK. A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- 8.Czapla L, Swigon D, Olson WK. Sequence-dependent effects in the cyclization of short DNA. J. Chem. Theor. Comp. 2006;2:685–695. doi: 10.1021/ct060025+. [DOI] [PubMed] [Google Scholar]
- 9.Tolstorukov MY, Colasanti AV, McCandlish D, Olson WK, Zhurkin VB. A novel ‘roll-and-slide’ mechanism of DNA folding in chromatin. Implications for nucleosome positioning. J. Mol. Biol. 2007;371:725–738. doi: 10.1016/j.jmb.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balasubramanian S, Xu F, Olson WK. DNA sequence-directed organization of chromatin: structure-based analysis of nucleosome-binding sequences. Biophys. J. 2009;96:2245–2260. doi: 10.1016/j.bpj.2008.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MySQL. Available at http://www.mysql.com.
- 12.IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN) Abbreviations and symbols for the description of conformations of polynucleotide chains. Eur. J. Biochem. 1983;131:9–15. doi: 10.1111/j.1432-1033.1983.tb07225.x. [DOI] [PubMed] [Google Scholar]
- 13.Altona C, Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J. Am. Chem. Soc. 1972;94:8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- 14.Xin Y, Olson WK. BPS: a database of RNA base-pair structures. Nucleic Acids Res. 2008;37:D83–D88. doi: 10.1093/nar/gkn676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickerson RE, Bansal M, Calladine CR, Diekmann S, Hunter WN, Kennard O, von Kitzing E, Lavery R, Nelson HCM, Olson WK, et al. Definitions and nomenclature of nucleic acid structure parameters. J. Mol. Biol. 1989;208:787–791. [Google Scholar]
- 16.Zhurkin VB, Lysov YP, Ivanov VI. Anisotropic flexibility of DNA and the nucleosomal structure. Nucleic Acids Res. 1979;6:1081–1096. doi: 10.1093/nar/6.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Hassan MA, Calladine CR. The assessment of the geometry of dinucleotide steps in double-helical DNA: a new local calculation scheme. J. Mol. Biol. 1995;251:648–664. doi: 10.1006/jmbi.1995.0462. [DOI] [PubMed] [Google Scholar]
- 18.Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinemann U, Lu X-J, Neidle S, Shakked Z, et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 19.Babcock MS, Pednault EPD, Olson WK. Nucleic acid structure analysis: mathematics for local Cartesian and helical structure parameters that are truly comparable between structures. J. Mol. Biol. 1994;237:125–156. doi: 10.1006/jmbi.1994.1213. [DOI] [PubMed] [Google Scholar]
- 20.Guzikevich-Guerstein G, Shakked Z. A novel form of the DNA double helix imposed on the TATA-box by the TATA-binding protein. Nat. Struct. Biol. 1995;3:32–37. doi: 10.1038/nsb0196-32. [DOI] [PubMed] [Google Scholar]
- 21.El Hassan MA, Calladine CR. Two distinct modes of protein-induced bending in DNA. J. Mol. Biol. 1998;282:331–343. doi: 10.1006/jmbi.1998.1994. [DOI] [PubMed] [Google Scholar]
- 22.CodeIgniter. Available at http://codeigniter.com.
- 23.Jmol. Available at http://www.jmol.org.
- 24.Olson WK, Colasanti AV, Czapla L, Zheng G. In: Coarse-Graining of Condensed Phase and Biomolecular Systems. Voth GA, editor. LLC: Taylor and Francis Group; 2008. pp. 205–223. [Google Scholar]
- 25.Goodsell DS, Kopka ML, Dickerson RE. Refinement of netropsin bound to DNA: bias and feedback in electron density map interpretation. Biochemistry. 1995;34:4983–4993. doi: 10.1021/bi00015a009. [DOI] [PubMed] [Google Scholar]