Abstract
‘Recoding’ is a term used to describe non-standard read-out of the genetic code, and encompasses such phenomena as programmed ribosomal frameshifting, stop codon readthrough, selenocysteine insertion and translational bypassing. Although only a small proportion of genes utilize recoding in protein synthesis, accurate annotation of ‘recoded’ genes lags far behind annotation of ‘standard’ genes. In order to address this issue, provide a service to researchers in the field, and offer training data for developers of gene-annotation software, we have gathered together known cases of recoding within the Recode database. Recode-2 is an improved and updated version of the database. It provides access to detailed information on genes known to utilize translational recoding and allows complex search queries, browsing of recoding data and enhanced visualization of annotated sequence elements. At present, the Recode-2 database stores information on approximately 1500 genes that are known to utilize recoding in their expression—a factor of approximately three increase over the previous version of the database. Recode-2 is available at http://recode.ucc.ie
INTRODUCTION
The term ‘translational recoding’ describes the utilization of non-standard decoding during protein synthesis and encompasses such processes as ribosomal frameshifting, codon redefinition, translational bypassing and StopGo (1–7). What is often considered as a decoding error—e.g. a frameshifting error or mistranslation of a particular codon—may occasionally benefit the organism by increasing its fitness and survival. In such instances the propensity for the decoding ‘error’ may be selected for during evolution, leading to the formation of a particular sequence context that elevates the frequency of the ‘error’. To discriminate such cases of programmed decoding ‘misbehaviour’ from promiscuous translational errors or translational noise, the term recoding is used. The position within an mRNA where a recoding event takes place is termed the ‘recoding site’. Sequence elements responsible for increasing the efficiency of recoding events are termed ‘recoding stimulatory signals’, and a minimal sequence fragment that allows recoding to take place at the natural efficiency (i.e. relative to the level of standard decoding at the recoding site) is termed a ‘recoding cassette’.
Recoding can benefit gene expression in a number of ways. It can regulate gene expression by being part of a sensor for particular cellular conditions. Prominent examples include ribosomal frameshifting in bacterial release factor 2 (RF2) and eukaryotic antizyme mRNAs. In both instances, ribosomal frameshifting is required for the production of the corresponding active full-length protein products. In the RF2 mRNA, the efficiency of frameshifting is negatively regulated by the cellular concentration of its product, RF2, providing an auto-regulatory circuit for its biosynthesis (8–10). In the antizyme mRNA, the efficiency of frameshifting is modulated by cellular levels of polyamines, whose concentration in turn is controlled by antizyme (11,12). Thus, this mechanism ensures the maintenance of antizyme production at the levels required to support physiologically appropriate concentrations of polyamines. Recoding can also be used for the diversification of protein products encoded by a single gene. An illustrative example is in bacterial dnaX mRNA, where frameshifting allows synthesis of two different protein subunits—sharing the same N-terminal part—from a single open reading frame (ORF) in its mRNA (13–15). A presumed constant ratio of frameshifting in dnaX ensures a fixed stoichiometric balance between these two subunits (16). This balance, then, is independent of the absolute levels of dnaX transcription and translational initiation on its mRNA. Similarly, in many viruses recoding is responsible for setting a ratio between protein products (such as those encoded by gag–pro–pol genes in retroviruses) produced from a single mRNA (17). Recoding also provides RNA viruses with a mechanism for the translation of downstream ORFs on polycistronic RNAs [other mechanisms include leaky scanning, shunting, reinitiation, IRESs and the production of subgenomic RNAs (18)] and may also be involved in global regulation mechanisms, such as mediating the switch between translation and replication on the same genomic RNA (19). Finally, recoding provides a way for the incorporation of non-standard amino acids—e.g. amino acids that share their codons with termination signals (the most prominent example of which is selenocysteine, encoded by UGA) (20–22). For further information on the diverse variety of recoding functions, see recent reviews (1,3,7,23,24).
Recoding cassettes may be composed of a variety of diverse sequence elements. For example, primary nucleotide sequences may promote re-arrangements of tRNA molecules relative to their codons in mRNA inside the ribosome or affect recognition of tRNAs or release factors in the ribosomal A-site. On the other hand, many recoding signals act in the form of RNA secondary structures, such as simple stem-loops, or more complex pseudoknots, kissing stem-loops and other structures that involve interactions between considerably distant RNA regions (19,25–28). Trans-acting RNA signals affecting ribosomal decoding through complementary interactions with ribosomal RNA (29–32), or through the nascent peptide acting within the ribosome exit tunnel (6,33,34), are also known. Some recoding events—such as selenocysteine insertion—require the presence of additional specialized machinery such as selenocysteine tRNAs, selenocysteine-specific translation factors and several other components of the selenocysteine biosynthesis and insertion pathway (20,35–37). Recent reviews on stimulatory signals involved in the modulation of recoding events and molecular mechanisms of recoding provide further details (7,25,27,38,39).
Despite considerable progress in the development of computational tools for the prediction of protein coding genes in sequenced genomes, the identification and annotation of recoded genes lags far behind. The hurdle lies not so much in the fact that recoded genes do not obey standard rules of genetic readout but, rather, in the considerable diversity of recoded genes and sequence elements responsible for recoding. Even among evolutionarily related genes, all utilizing recoding, the diversity of recoding signals can be considerable. An extreme example is when orthologous genes utilize recoding at different stages of gene expression to achieve the same goal. An example is in dnaX, where ribosomal frameshifting is employed by enterobacteria, but transcriptional slippage is used in Thermus thermophilus (40). A similar situation occurs in bacterial insertion sequence (IS) elements, where a certain group of IS elements utilizes transcriptional slippage to produce ORFA–ORFB fusions, while many other IS elements utilize ribosomal frameshifting for the same purpose (41). The diversity of recoding functions, combined with the wide spectrum of unrelated sequence elements involved in recoding, makes the design of a uniform model of recoding intractable. Nonetheless, in recent years, we have witnessed the development of specialized models and computational tools for the identification of particular subsets of recoding cassettes, or tools that are specific to recoding events in particular groups of homologous genes (42–45).
These developments, at least partially, were facilitated by the availability of a compiled dataset of known recoded genes collected together in the Recode database (http://recode.genetics.utah.edu), which was initially launched 9 years ago (46,47). To facilitate further development of computational tools for the prediction of recoded genes in the ever faster growing body of sequence data, as well as to provide bench researchers with up-to-date information on recoding, an efficient means of Recode database population and annotation are now required. In this article, we describe the incarnation of the database, Recode-2. The major advances of Recode-2 (hosted in a new location http://recode.ucc.ie) over previous versions include a new web design allowing enhanced visualization of stimulatory signals, a uniform RecodeML format for the annotation of recoded genes, and a significantly larger number of entries—including many recently identified cases—that altogether have more than doubled the size of the database since its last published update.
DATABASE ORGANIZATION AND USAGE
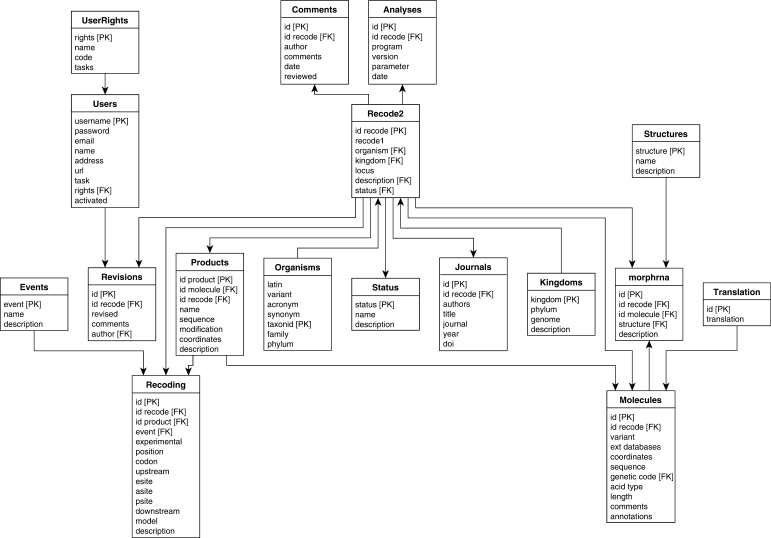
The data are stored in a local PostgreSQL database that is queried by PHP scripts embedded in the web interface. The schema of the PostgreSQL database is shown in Figure 1. The database stores information on individual genes that utilize recoding, the mechanisms and stimulatory signals involved, and references to the original literature sources that describe the recoding events. In order to facilitate the uniform annotation of recoding events, we have designed an XML-based format for the annotation of recoded genes, RecodeML. The document type definition for RecodeML is available at the Recode-2 web site at http://recode.ucc.ie/dtd The extensibility of the RecodeML format will allow incorporation of new annotation, if required, for newly discovered types of recoding, and the associated features, as they are being discovered. The database handles batch importation of properly designed RecodeML entries into the PostgreSQL database, thus facilitating rapid population of the database with new data.
Figure 1.
Entity-Relationship diagram for the Recode-2 database. The database schematic shows the relationships between the database tables. Each Recode-2 entry is composed of one entry in the Recode-2 table and a variable number of entries in related tables. PK (Primary Key) indicates the row selected as the unique identifier for a table. FK (Foreign Key) indicates rows of a table whose values match those in the primary key of a related table. (This ensures the constancy of the database.) The arrows show the connection relationships between the various tables (PK to FK).
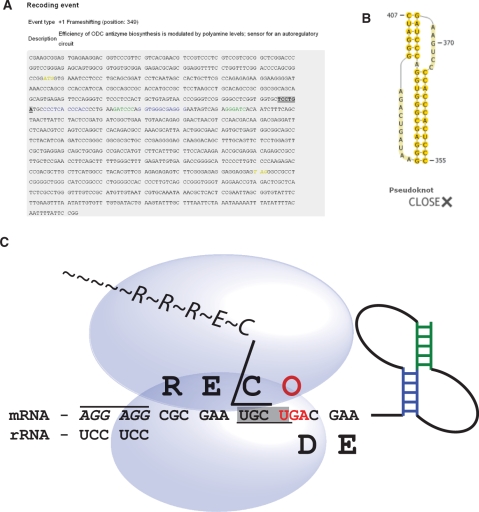
The data in the database may be explored in two ways. They may be browsed by one of the three categories: kingdom (archaea, bacteria, eukaryotes and viruses), organism and type of recoding. The data may also be searched directly by key words that can be inserted into the search field. Searches that use regular expressions are allowed. The output of a database search is a list of Recode-2 entries in a short format that includes organism name, kingdom, genus, type of recoding event, status of the entry in the database and a link to the full database entry. The full description includes the following additional information: (i) the common name of the gene and the validation status of the recoding event; (ii) the organism description, giving the organism name and a link to the NCBI Taxonomy Browser (48); (iii) the sequence description, giving the Genbank (49) accession numbers for matching sequences (with links to Genbank) and links to detailed annotations of the sequences and to diagrams of RNA secondary structures involved in stimulation of the recoding event; (iv) information on the protein sequence generated as a result of recoding; (v) comments on the function of the recoding event and any additional notes and (vi) references to relevant literature (with links to corresponding abstracts in PubMed). The detailed sequence annotation appears in the form of text decorations that are described in the Help page of the database and are also illustrated within the Recode-1 logo itself (which can be used for rapid intuitive decoding of the text decorations and their associations with the mechanistic ways by which different sequences affect ribosome functions). To generate RNA secondary structure diagrams, PseudoViewer3 (50) is used, since it can handle complex pseudoknot RNA structures correctly. Figure 2 shows an example of sequence annotation for the human oaz1 gene, alongside a diagram of a stimulatory RNA secondary structure, and the Recode-1 logo.
Figure 2.
Screenshots from the Recode-2 database for the entry describing +1 frameshifting in human antizyme 1. (A) Annotated sequence of the human antizyme 1 gene. Colouring and formatting of the text highlights functional elements according to a scheme described on the Help page of the Recode-2 database and within the Recode-1 logo. (B) Structure of the stimulatory RNA pseudoknot. (C) The Recode1 logo.
NOVEL DATA
Unlike Recode-1, where all data on recoding events were introduced manually, Recode-2 also utilizes automated identification of recoding events by the recently developed computer programs ARFA (43) and OAF (44), that are able to identify and annotate +1 frameshifting events in mRNAs of bacterial RF2s and eukaryotic antizyme (OAZs), respectively. However, a significant source of recoding events remains to be serendipitous discoveries by experimental studies that sometimes are complemented by more systematic studies of large groups of similar genes (51,52). Therefore, a large proportion of new data are still populated manually or semi-manually. To ease manual population of recoding events, a special form has been designed that is available in the database upon user registration. User registration needs to be approved by one of the database contributors. The novel data in the database include 249 RF2 mRNAs identified by ARFA, 152 events identified by OAF, 200 new selenoprotein genes (53–56) and ∼200 new viral annotations (57) including the newly discovered frameshift cassettes in potyviruses (58), alphaviruses (59) and the Japanese encephalitis group of flaviviruses (60).
FUTURE DEVELOPMENT
The database will expand in accordance with the growth of available sequence information that will be scanned by one of the existing programs for recode annotation. We also plan to continue developing tools for the automatic identification of recoding events from nucleotide sequences. As the field grows and the number of recoded genes progressively increases, it becomes harder to extract data from the relevant literature and a number of novel recoded genes may escape the database. Therefore, we encourage users and researchers in the field to submit their data directly to the Recode-2 database. We are also willing to provide help with the analysis of potential new recoding events.
FUNDING
Science Foundation Ireland (SFI) grants (to P.V.B. and J.F.A.); National Institutes of Health grants (to J.F.A. and V.N.G.). Funding for open access charge: Science Foundation Ireland.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to express our appreciation to the colleagues who have contributed data for the previous versions of the database.
REFERENCES
- 1.Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 2.Farabaugh PJ. Programmed translational frameshifting. Annu. Rev. Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 3.Baranov PV, Gesteland RF, Atkins JF. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- 4.Dinman JD. Programmed ribosomal frameshifting goes beyond viruses: organisms from all three kingdoms use frameshifting to regulate gene expression, perhaps signaling a paradigm shift. Microbe. 2006;1:521–527. doi: 10.1128/microbe.1.521.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkins JF, Wills NM, Loughran G, Wu CY, Parsawar K, Ryan MD, Wang CH, Nelson CC. A case for “StopGo”: reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go) RNA. 2007;13:803–810. doi: 10.1261/rna.487907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herr AJ, Atkins JF, Gesteland RF. Coupling of open reading frames by translational bypassing. Annu. Rev. Biochem. 2000;69:343–372. doi: 10.1146/annurev.biochem.69.1.343. [DOI] [PubMed] [Google Scholar]
- 7.Atkins JF, Gesteland RF. Recoding: expansion of decoding rules enriches gene expression. New York: Springer; 2010. [Google Scholar]
- 8.Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 9.Craigen WJ, Caskey CT. The function, structure and regulation of E. coli peptide chain release factors. Biochimie. 1987;69:1031–1041. doi: 10.1016/0300-9084(87)90003-4. [DOI] [PubMed] [Google Scholar]
- 10.Baranov PV, Gesteland RF, Atkins JF. Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 2002;3:373–377. doi: 10.1093/embo-reports/kvf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov IP, Atkins JF. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flower AM, McHenry CS. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 1990;87:3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuchihashi Z, Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl Acad. Sci. USA. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blinkowa AL, Walker JR. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 1990;18:1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen B, Gesteland RF, Atkins JF. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50% J. Mol. Biol. 1997;271:47–60. doi: 10.1006/jmbi.1997.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brierley I, Dos Ramos FJ. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006;119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiebeauld O, Pooggin MM, Ryabova LA. Alternative translation strategies in plant viruses. Plant Viruses. 2007;1:1–20. [Google Scholar]
- 19.Miller WA, White KA. Long-distance RNA–RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Squires JE, Berry MJ. Eukaryotic selenoprotein synthesis: mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life. 2008;60:232–235. doi: 10.1002/iub.38. [DOI] [PubMed] [Google Scholar]
- 21.Allmang C, Krol A. Selenoprotein synthesis: UGA does not end the story. Biochimie. 2006;88:1561–1571. doi: 10.1016/j.biochi.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 22.Hatfield DL, Berry MJ, Gladyshev VN. Selenium: Its Molecular Biology and Role in Human Health. New York: Springer; 2006. [Google Scholar]
- 23.Baranov PV, Fayet O, Hendrix RW, Atkins JF. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Plant EP, Dinman JD. The role of programmed-1 ribosomal frameshifting in coronavirus propagation. Front Biosci. 2008;13:4873–4881. doi: 10.2741/3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giedroc DP, Theimer CA, Nixon PL. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brierley I, Gilbert RJ, Pennell S. RNA pseudoknots and the regulation of protein synthesis. Biochem. Soc. Trans. 2008;36:684–689. doi: 10.1042/BST0360684. [DOI] [PubMed] [Google Scholar]
- 28.Barry JK, Miller WA. A -1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl Acad. Sci. USA. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss RB, Dunn DM, Atkins JF, Gesteland RF. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- 30.Larsen B, Peden J, Matsufuji S, Matsufuji T, Brady K, Maldonado R, Wills NM, Fayet O, Atkins JF, Gesteland RF. Upstream stimulators for recoding. Biochem. Cell Biol. 1995;73:1123–1129. doi: 10.1139/o95-121. [DOI] [PubMed] [Google Scholar]
- 31.Atkins JF, Baranov PV, Fayet O, Herr AJ, Howard MT, Ivanov IP, Matsufuji S, Miller WA, Moore B, Prere MF, et al. Overriding standard decoding: implications of recoding for ribosome function and enrichment of gene expression. Cold Spring Harb. Symp. Quant. Biol. 2001;66:217–232. doi: 10.1101/sqb.2001.66.217. [DOI] [PubMed] [Google Scholar]
- 32.Curran JF, Yarus M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J. Mol. Biol. 1988;203:75–83. doi: 10.1016/0022-2836(88)90092-7. [DOI] [PubMed] [Google Scholar]
- 33.Wills NM, O'C;onnor M, Nelson CC, Rettberg CC, Huang WM, Gesteland RF, Atkins JF. Translational bypassing without peptidyl-tRNA anticodon scanning of coding gap mRNA. EMBO J. 2008;27:2533–2544. doi: 10.1038/emboj.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss RB, Huang WM, Dunn DM. A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell. 1990;62:117–126. doi: 10.1016/0092-8674(90)90245-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lescure A, Fagegaltier D, Carbon P, Krol A. Protein factors mediating selenoprotein synthesis. Curr. Protein Pept. Sci. 2002;3:143–151. doi: 10.2174/1389203023380783. [DOI] [PubMed] [Google Scholar]
- 36.Walczak R, Hubert N, Carbon P, Krol A. Solution structure of SECIS, the mRNA element required for eukaryotic selenocysteine insertion–interaction studies with the SECIS-binding protein SBP. Biomed. Environ. Sci. 1997;10:177–181. [PubMed] [Google Scholar]
- 37.Commans S, Bock A. Selenocysteine inserting tRNAs: an overview. FEMS Microbiol. Rev. 1999;23:335–351. doi: 10.1111/j.1574-6976.1999.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 38.Baranov PV, Gesteland RF, Atkins JF. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA. 2004;10:221–230. doi: 10.1261/rna.5122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao PY, Gupta P, Petrov AN, Dinman JD, Lee KH. A new kinetic model reveals the synergistic effect of E-, P- and A-sites on +1 ribosomal frameshifting. Nucleic Acids Res. 2008;36:2619–2629. doi: 10.1093/nar/gkn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larsen B, Wills NM, Nelson C, Atkins JF, Gesteland RF. Nonlinearity in genetic decoding: homologous DNA replicase genes use alternatives of transcriptional slippage or translational frameshifting. Proc. Natl Acad. Sci. USA. 2000;97:1683–1688. doi: 10.1073/pnas.97.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baranov PV, Hammer AW, Zhou J, Gesteland RF, Atkins JF. Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 2005;6:R25. doi: 10.1186/gb-2005-6-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Theis C, Reeder J, Giegerich R. KnotInFrame: prediction of -1 ribosomal frameshift events. Nucleic Acids Res. 2008;36:6013–6020. doi: 10.1093/nar/gkn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bekaert M, Atkins JF, Baranov PV. ARFA: a program for annotating bacterial release factor genes, including prediction of programmed ribosomal frameshifting. Bioinformatics. 2006;22:2463–2465. doi: 10.1093/bioinformatics/btl430. [DOI] [PubMed] [Google Scholar]
- 44.Bekaert M, Ivanov IP, Atkins JF, Baranov PV. Ornithine decarboxylase antizyme finder (OAF): fast and reliable detection of antizymes with frameshifts in mRNAs. BMC Bioinformatics. 2008;9:178. doi: 10.1186/1471-2105-9-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon S, Byun Y, Kim HJ, Jeong S, Han K. Predicting genes expressed via -1 and +1 frameshifts. Nucleic Acids Res. 2004;32:4884–4892. doi: 10.1093/nar/gkh829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baranov PV, Gurvich OL, Fayet O, Prere MF, Miller WA, Gesteland RF, Atkins JF, Giddings MC. RECODE: a database of frameshifting, bypassing and codon redefinition utilized for gene expression. Nucleic Acids Res. 2001;29:264–267. doi: 10.1093/nar/29.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baranov PV, Gurvich OL, Hammer AW, Gesteland RF, Atkins JF. Recode 2003. Nucleic Acids Res. 2003;31:87–89. doi: 10.1093/nar/gkg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Byun Y, Han K. PseudoViewer3: generating planar drawings of large-scale RNA structures with pseudoknots. Bioinformatics. 2009;25:1435–1437. doi: 10.1093/bioinformatics/btp252. [DOI] [PubMed] [Google Scholar]
- 51.Gurvich OL, Baranov PV, Zhou J, Hammer AW, Gesteland RF, Atkins JF. Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. EMBO J. 2003;22:5941–5950. doi: 10.1093/emboj/cdg561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu J, Hendrix RW, Duda RL. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell. 2004;16:11–21. doi: 10.1016/j.molcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Gladyshev VN. Comparative genomics of trace elements: emerging dynamic view of trace element utilization and function. Chem. Rev. 2009 doi: 10.1021/cr800557s. in press. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Romero H, Salinas G, Gladyshev VN. Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006;7:R94. doi: 10.1186/gb-2006-7-10-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Gladyshev VN. Trends in selenium utilization in marine microbial world revealed through the analysis of the global ocean sampling (GOS) project. PLoS Genet. 2008;4:e1000095. doi: 10.1371/journal.pgen.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim HY, Zhang Y, Lee BC, Kim JR, Gladyshev VN. The selenoproteome of Clostridium sp. OhILAs: characterization of anaerobic bacterial selenoprotein methionine sulfoxide reductase A. Proteins. 2009;74:1008–1017. doi: 10.1002/prot.22212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bekaert M, Rousset JP. An extended signal involved in eukaryotic -1 frameshifting operates through modification of the E site tRNA. Mol. Cell. 2005;17:61–68. doi: 10.1016/j.molcel.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chung BY, Miller WA, Atkins JF, Firth AE. An overlapping essential gene in the Potyviridae. Proc. Natl Acad. Sci. USA. 2008;105:5897–5902. doi: 10.1073/pnas.0800468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Firth AE, Chung BY, Fleeton MN, Atkins JF. Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma. Virol. J. 2008;5:108. doi: 10.1186/1743-422X-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Firth AE, Atkins JF. A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1' may derive from ribosomal frameshifting. Virol. J. 2009;6:14. doi: 10.1186/1743-422X-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]