Abstract
The NCBI BioSystems database, found at http://www.ncbi.nlm.nih.gov/biosystems/, centralizes and cross-links existing biological systems databases, increasing their utility and target audience by integrating their pathways and systems into NCBI resources. This integration allows users of NCBI’s Entrez databases to quickly categorize proteins, genes and small molecules by metabolic pathway, disease state or other BioSystem type, without requiring time-consuming inference of biological relationships from the literature or multiple experimental datasets.
INTRODUCTION
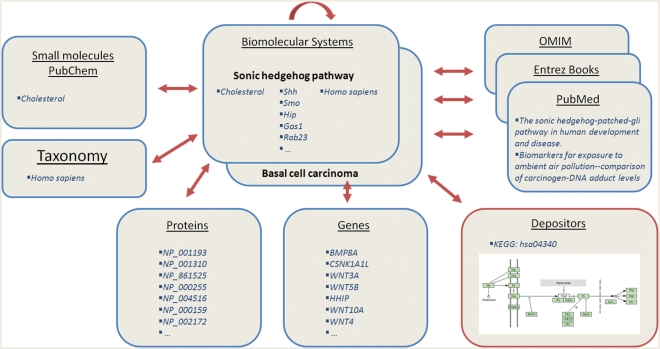
Biological molecular databases often contain relationships between records based on computational inference of similarity, such as links between sequences deemed homologous in protein and nucleotide databases. Less frequently do they explicitly log relationships between records that are experimentally derived, such as the genes interacting in a biological pathway, even though knowledge of these relationships is crucial for understanding living systems and for performing biological research. Fortunately, a considerable number of resources have been created to address this issue: the Pathguide (1) resource lists nearly 300 pathway resources alone, including KEGG (2), Reactome (3), PID (4), PharmGKB (5), GenMAPP (6), Biocyc (7) and many others. While there is some degree of overlap between such resources, there may be significant numbers of unique records available from many of the underlying datasets. However, because of the diverse history of these databases and resources, integration with commonly used molecular database resources, such as NCBI’s Entrez search engine, is done on a case-by-base basis. To address this issue, we have created the NCBI BioSystems database that functions as a clearinghouse for these databases by integrating their data into the existing NCBI Entrez databases (8), such as Gene, Protein, PubMed and PubChem, and linking back to the original database web site for more detailed information and analysis (Figure 1). Centralizing and linking the existing biosystems databases potentially increase their usefulness by integrating their pathways and systems into a resource that is accessed by a significant number of scientists. It also enables users to quickly find and categorize proteins, genes and small molecules by pathway, disease state, etc., instead of requiring time-consuming inference of biological relationships from other evidence, e.g. by examining a 3D structure.
Figure 1.
A schematic representation of the integration of the BioSystems database with various NCBI resources and with resources publically available from the depositor.
OVERVIEW OF CONTENT
A BioSystem record is defined as a biologically related list of gene, protein and small molecule identifiers, along with the characterization of interactions, citations and other annotations, where none of these items are mandatory. This definition is not limited to metabolic- or signaling pathways: for example, a BioSystems disease record may contain susceptibility genes, biomarkers and drugs used for treatment.
The BioSystems database is archival and each BioSystem record receives a unique identifier known as a bsid that is intended to remain constant over the lifetime of the record. Each new version of a BioSystem record is assigned a version number.
Presently, NCBI BioSystems contains pathways from KEGG (2), Human Reactome (3) and EcoCyc (9) for a total of about 100 000 BioSystem records. These BioSystems records link to over 2 million protein records, nearly 900 000 gene records and several thousands PubChem records.
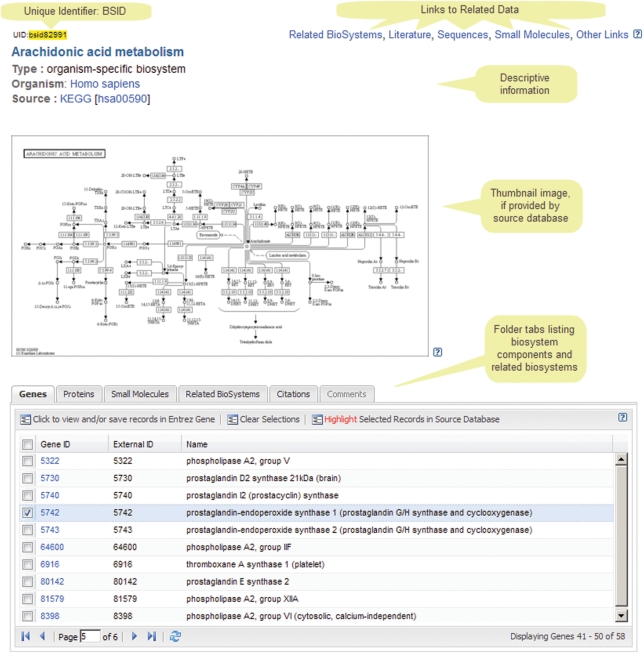
An example record, shown in Figure 2, describes the COX portion of the human arachidonic acid metabolism pathway, which metabolizes lipids into prostaglandins that are involved in a host of regulatory mechanisms via binding to and activating G protein-coupled receptors. This pathway has an important role in pain and inflammation. Specifically, the protein encoded by human PTGS1 gene is involved in the conversion of prostaglandin PGG2 into inflammation-causing prostaglandin PGH2. Aspirin has been shown to bind to the PTGS1 gene product (prostaglandin-endoperoxide synthase 1), blocking that enzyme’s ability to produce PGH2 and thereby reducing pain and inflammation. The NCBI BioSystems record lists these genes, their associated proteins and the small molecules involved in the pathway. The BioSystems records also contain annotations such as taxonomy, description, pathway images and citations. Finally, links to and from other NCBI Entrez databases are listed, including links between BioSystems records. Links between BioSystems records are specified by the depositor and also generated computationally for BioSystems that list overlapping sets of proteins.
Figure 2.
An example BioSystems record display for the COX portion of the arachidonic acid metabolism pathway, which metabolizes lipids that are commonly found in the cell membrane into prostaglandins. The display includes a thumbnail image, links back to the depositor’s web site, and lists of the molecular identifiers and annotations associated with the pathway.
Currently, we distinguish between two major record types, organism-specific biosystems and conserved biosystems. Organism-specific biosystems correspond to particular instances of a biological system, such as the arachidonic acid pathway in human. Conserved biosystems are canonical biosystems that are used to group together orthologous, organism-specific biosystems. Currently, these records are derived from reference pathways in the KEGG database.
DATA PROCESSING AND INTEGRATION
Two major issues were addressed in the creation of the BioSystems database: loading data from disparate data sources and integration of the data into the current NCBI Entrez database infrastructure.
Publicly available biosystems databases organize their data in significantly different ways, including the use of a variety of molecular identifiers and formatting their data in database-specific schemas. Even when databases support well-established data standards such as BioPAX (10) or SBML (11), there are situations where the standards may not provide for encoding of some data, such as pathway graphical images, or allow ambiguity that makes automated import more difficult, such as not explicitly enumerating sequence source database names in sequence identifiers. To avoid these issues when depositing data into the NCBI BioSystems database, we created the Really Simple System Markup XML data specification. The specification is intentionally trivial in structure and encourages unambiguous specification of molecular identifiers.
Integration of the resulting deposition into the NCBI Entrez system requires multiple data processing steps. For example, one depositor may prefer giving gene ids, while another may prefer giving Uniprot accessions. In both cases, the depositor may wish that we link to all applicable gene ids and all identical sequence accessions to maximize the amount of BioSystem annotations provided to NCBI users. The following is a list of the NCBI resources that are linked to along with the methods currently used. All of the links are updated, at minimum, on a weekly basis using the current version of the database being linked to.
Proteins
Protein GI numbers present in the source record are parsed out, and links are then established directly to the corresponding sequence records in the Entrez Protein database. If the source record contains protein accessions, the current GI number for each accession is determined and a link to the corresponding protein sequence record is made using the derived GI number. In addition, the set of links to protein sequences is expanded in the following ways: (i) if any GI numbers are for RefSeq records, links to corresponding UniProt/Swiss-Prot (12) records are also made; (ii) if any other record(s) in the Entrez Protein database contains an identical sequence to the one present in the cited GI and also share the same NCBI Taxonomy ID (TaxID), links to those identical sequence records are established as well; and (iii) if the record is linked to GeneIDs, then all proteins linked to those GeneIDs are linked to.
Genes
GeneIDs present in the source record are parsed out and links are then established to the corresponding records in the Entrez Gene database. Links are also established to Gene IDs that correspond to the protein sequence GI numbers mentioned above; for example, if one of those protein GIs is cited directly in a Gene record, a link to that Gene record is made.
Small molecules
Records from source databases are parsed for small molecule identification numbers, including PubChem (13), Compound IDs (CIDs), PubChem Substance IDs (SIDs) and external registry names. The types of links that are made depend upon the type of identifiers that were found: If SIDs are present in the source record, links are established to the corresponding PubChem Substance records and to associated CIDs in PubChem Compound. If CIDs are present in the source record, links to the corresponding PubChem Compound records are made (however, the links are not extended to associated PubChem Substances). If external registry names are present, those identifiers are mapped to the corresponding SIDs and links are made to those records in PubChem Substance as well as to associated CIDs in PubChem Compound.
Literature
If the source record includes PubMed identifiers (PMIDs) for journal articles about the biosystem, the PMIDs are parsed and links are established to the corresponding records in the PubMed database.
Taxonomy
Depositors provide the Taxonomy ID (TaxID) of the source organism for organism-specific biosystems. These TaxIDs are parsed and links to the corresponding information in the NCBI Taxonomy database are then established. Taxonomic information is not extracted from conserved biosystems.
BioSystems
A depositor can explicitly link together BioSystems, such as from one whose product is the substrate of another.
Using these links and other links available in the Entrez search system, a series of indirect links are calculated, including:
Bioassays: bioactivity screens of small molecules where the target of the screen is a protein whose sequences are also found in BioSystems records.
3D protein structures: 3D protein structures whose corresponding sequences are also found in BioSystems records.
Functionally related sequences: calculated by links to protein sequences that have specific hits to Conserved Domains and also to sequences contained in HomoloGene and Protein Cluster groups.
Genetic phenotypes: Mendelian disorders and genes listed in the Online Mendelian Inheritance in Man database, calculated by using links to Entrez Gene.
Related BioSystems: two or more biosystem records are linked together as related if the biosystems share at least one identical protein sequence from the same source organism. The identical sequence and same organism requirements tend to relate records from the same data source, as different data sources can use different strains and slightly different sequences for the same enzyme. This issue can be addressed in future by using gene records for the link calculation and also matching organisms at the species level.
AVAILABILITY
The BioSystems database is searchable by keyword on the web using the NCBI Entrez system. Figure 2 shows what a typical record displayed in this system might look like. When available, the record comes with a graphical representation of the BioSystem, and, below that, tabbed lists of associated genes, proteins, small molecules, citations and other annotations. The tabbed lists allow for sorting, selection and filtering and, when supported by the depositor, selected proteins, genes and small molecules can be highlighted in graphical representations of the BioSystem by using web services provided by the depositor’s site.
The data and most of the links generated in the steps outlined above are available for download at ftp://ftp.ncbi.nih.gov/pub/biosystems/.
Programmatic access is available via the NCBI Entrez programming utilities (eutils) as described at http://eutils.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html.
The database is currently updated on a weekly basis and incorporates any new or changed data from data sources received in the previous week. The frequency of updates from particular data sources is determined by the data source. For example, KEGG sends weekly updates.
FUTURE DIRECTIONS
To aid discoverability, we plan further the integration of the NCBI BioSystems database with other components of NCBI’s Entrez system. This might include, for example, the display of relevant BioSystems information in Entrez Gene, Protein and PubChem small molecule records.
For analysis of data on a large scale, such as obtained via high-throughput experimentation, we anticipate the development of services that facilitate summary views of such data characterized by biosystems. For example, this might include an ordered list of the BioSystems most represented in a high-throughput biological assay.
Finally, we anticipate incorporating additional datasets to further increase the number of unique biosystems in our databases.
FUNDING
Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS. Funding for open access charge: Intramural Research Program of the National Library of Medicine at the National Institutes of Health/DHHS.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the authors of the KEGG, Reactome and BioCyc databases. We also thank the NCBI Information Engineering Branch for continuing assistance with software development.
REFERENCES
- 1.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, Hewett M, Lin Z, Liu Y, Liu S, Oliver DE, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1:167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 6.Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahren D, Tsoka S, Darzentas N, Kunin V, Lopez-Bigas N. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luciano JS. PAX of mind for pathway researchers. Drug Discov. Today. 2005;10:937–942. doi: 10.1016/S1359-6446(05)03501-4. [DOI] [PubMed] [Google Scholar]
- 11.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden nA, et al. The Systems Biology Markup Language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 12.The UniProt Consortium The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]