Abstract
DataBase of Transcription Start Sites (DBTSS) is a database which contains precise positional information for transcription start sites (TSSs) of eukaryotic mRNAs. In this update, we included 330 million new tags generated by massively sequencing the 5′-end of oligo-cap selected cDNAs in humans and mice. The tags were collected from normal fetal or adult human tissues, including brain, thymus, liver, kidney and heart, from 6 human cell lines in 21 diverse growth conditions as well as from mouse NIH3T3 cell line: altogether 31 different cell types or culture conditions are represented. This unprecedented increase in depth of data now allows DBTSS to faithfully represent the dynamically changing landscape of TSSs in different cell types and conditions, during development and in the course of evolution. Differential usage of alternative 5′-ends across cell types and conditions can be viewed in a series of new interfaces. Promoter sequence information is now displayed in a comparative genomics viewer where evolutionary turnover of the TSSs can be evaluated. DBTSS can be accessed at http://dbtss.hgc.jp/.
INTRODUCTION
Precise positional information on the transcription start sites (TSSs) and their expression levels are key for identifying the putative upstream promoter regions and understanding transcriptional regulation of the genes. For this purpose, we have constructed DBTSS, a DataBase of Transcription Start Sites (1). DBTSS is based on our unique data which are experimentally validated by 5′-end sequencing of full-length cDNA libraries constructed using our oligo-capping method (2). Recently, in order to facilitate the data collection, we developed a new method, which we named TSS Seq (3). This method combines the oligo-capping and the massively parallel sequencing technology (4), so that tens of millions of TSSs data can be generated from a single assay.
In the previous update, we have published 20 million transcription start tag data collected from human MCF7 (Human breast adenocarcinoma) and HEK293 (Human embryonic kidney) cells (1). A similar approach using the deep CAGE method (5) on human THP1 (Human acute monocytic leukemia) cells lead the RIKEN FANTOM4 consortium to add 6 million 5′-end tags to their database (6). Through in-depth analysis of the TSSs in particular cell types, it has become gradually clear that a large number of human genes contain multiple (alternative) promoters (7,8) and each mammalian cell seems to utilize its own set of promoters (9). Therefore, a simple catalogue of the promoters, as often provided in the pre-existing databases, cannot represent a global view of transcriptional regulation in human genes, which is highly diversified and changes dynamically depending on cellular circumstances. For this purpose, it is essential to collect TSS data from a wider collection of cell types in diverse cellular environments. An appropriate interface is also indispensable to represent the TSS collected from different data points in an integrative manner. In this update, DBTSS includes about 300 million TSS tags collected from 31 different TSS Seq libraries, each of which contains ∼10 million TSS tags. The TSS data sets from each of the TSS Seq libraries were interconnected in our new interface, so that users can empirically understand the differential usage of the promoters. Here, we describe the update of our DBTSS, which enables, for the first time, to illustrate the dynamic nature of the mammalian gene promoters.
NEW FEATURES
Statistics of the newly included TSS data
In this update, we have included a total of 330 533 354 new 36-bp-single-end-read TSS tags. These tags were collected from a series of oligp-capped libraries constructed from eight kinds of human normal tissues (brain, kidney, heart, fetal brain, fetal kidney, fetal heart, fetal thymus and fetal liver) and six cultured cell lines (colon cancer DLD1, B lymphocyte Ramos, bronchial epithelial cells BEAS2B, embryonic kidney HEK293, breast adenocarcinoma MCF7 and fetal lung TIG3 in humans) and fibroblast NIH3T3 cells in mice; for details on the origin of the cells, see http://dbtss.hgc.jp/cgi-bin/cell_type.cgi. We constructed the 5′-end libraries using six cell types cultured in different conditions, such as hypoxia or normoxia, and with or without IL4 treatment. Altogether, the current DBTSS includes 31 different cell types or culture conditions, each containing ∼10 million TSS tags (Table 1). Accession numbers for each dataset are given in http://dbtss.hgc.jp/cgi-bin/accession.cgi. Details of the experimental procedures are also described in http://dbtss.hgc.jp/docs/protocol_solexa.html.
Table 1.
Statistics of the new TSS Seq data
| Panel A | |||||||
|---|---|---|---|---|---|---|---|
| Sample name | Cell type | Condition | Time course | Tag count | |||
| DLD1 (Hypoxia with non-tagged RNAi) | Fibroblast | 1% O2 | 24 h | 7 723 359 | |||
| DLD1 (Hypoxia with HIF1A RNAi) | Fibroblast | 1% O2 | 24 h | 7 727 105 | |||
| DLD1 (Normoxia with HIF1A RNAi) | Fibroblast | 21% O2 | 24 h | 7 410 902 | |||
| DLD1 (Hypoxia with HIF2A RNAi) | Fibroblast | 1% O2 | 24 h | 8 737 554 | |||
| DLD1 (Normoxia with non-targetedRNAi) | Fibroblast | 21% O2 | 24 h | 8 644 835 | |||
| DLD1 (Normoxia with HIF2A RNAi) | Fibroblast | 21% O2 | 24 h | 8 353 702 | |||
| Beas2B overexpress STAT6 IL4+ | Bcell | IL4 | 4 h | 22 954 017 | |||
| Beas2B overexpress STAT6 IL4− | Bcell | 21 127 774 | |||||
| Beas2B parent IL4+ | Bcell | IL4 | 4 h | 15 166 848 | |||
| Beas2B parent IL4− | Bcell | 11 628 747 | |||||
| Beas2B stat6 siRNA− IL4+ | Bcell | IL4 | 4 h | 8 243 100 | |||
| Beas2B stat6 siRNA− IL4− | Bcell | 7 857 509 | |||||
| Beas2B stat6 siRNA+ IL4+ | Bcell | IL4 | 4 h | 5 879 777 | |||
| Beas2B stat6 siRNA+ IL4− | Bcell | 5 931 745 | |||||
| Ramos IL4+ | Bcell | IL4 | 4 h | 15 268 493 | |||
| Ramos IL4− | Bcell | 15 759 413 | |||||
| MCF7 O2 1% | Breast adenocarcinoma | 1% O2 | 24 h | 7 531 326 | |||
| MCF7 O2 21% | Breast adenocarcinoma | 21% O2 | 24 h | 13 609 932 | |||
| TIG O2 1% | Fetal lung | 1% O2 | 24 h | 8 848 737 | |||
| TIG O2 21% | Fetal lung | 21% O2 | 24 h | 9 235 808 | |||
| 293 O2 1% | Embryonic kidney | 1% O2 | 24 h | 10 590 128 | |||
| 293 O2 21% | Embryonic kidney | 21% O2 | 24 h | 8 162 101 | |||
| Fetal Heart | Normal fetal tissues | 10 182 282 | |||||
| Fetal Kidney | Normal fetal tissues | 8 424 482 | |||||
| Fetal Liver | Normal fetal tissues | 4 741 889 | |||||
| Fetal Thymus | Normal fetal tissues | 7 122 556 | |||||
| Fetal Brain | Normal fetal tissues | 11 285 710 | |||||
| Brain | Normal adult tissues | 11 561 960 | |||||
| Heart | Normal adult tissues | 9 378 901 | |||||
| Kidney | Normal adult tissues | 11 196 359 | |||||
| Mouse 3T3 | Fibroblast | 20 246 303 | |||||
| Total | 330 533 354 | ||||||
| Panel: B | ||||||||
|---|---|---|---|---|---|---|---|---|
| Average 5′-end clusters per cell | Average number of TSCs > 5ppm per cell | Number of represented genes (coverage against total RefSeq genes) | ||||||
| RefSeq region | 105 134 | 6972 | 17879/18001 (99%) | |||||
| Intergenic region | 40 537 | 1083 | ND | |||||
The TSS tags in every dataset were clustered into 500 bp-bins to separate transcription start clusters (TSCs), each of which may represent independent promoters [also see Ref. (3) for further details]. 5′-end clusters were further split according to whether they mapped in the vicinity of a RefSeq gene (from −50 Kb upstream from the 5′-end of a RefSeq transcript to the 3′-end of it) or further than 50 Kb away, in what we call an intergenic region. As summarized in Table 1, on average, there were ∼100 000 RefSeq transcription start clusters and 40 000 intergenic ones per cell and per culture condition. In spite of the generally large number of 5′-end clusters consistent with previous observations from ourselves (3) and others (5), most of the clusters were composed of one or two TSSs. The TSCs having significant expression levels, which may be prioritized for further biological functional characterizations, were relatively rare. The number of TSCs having expression levels of > 5 ppm (part per million tags; note that 1 ppm corresponds to 1 copy per cell, assuming every cell contains 1 million mRNA copies) is summarized in Table 1. Detailed statistics on every TSS Seq sub-dataset are shown in http://dbtss.hgc.jp/cgi-bin/cell_type.c. All data can be downloaded on our download site (ftp://ftp.hgc.jp/pub/hgc/db/dbtss/dbtss_ver7/).
TSS dynamics viewer
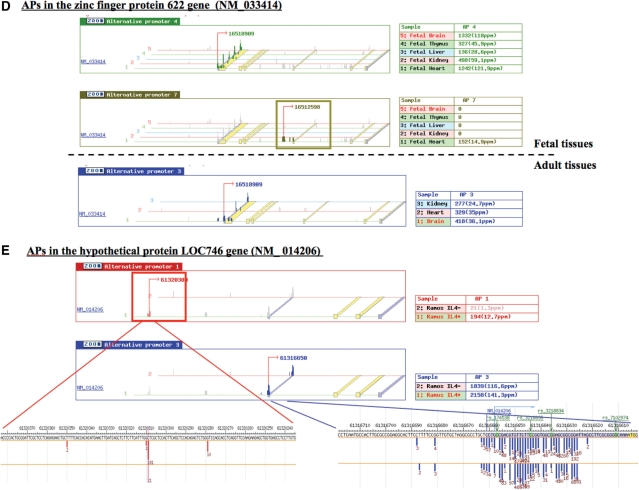
As the expanded DBTSS data contains hundreds of millions of TSS data collected from dozens of different cell types in diverse culture conditions, it is essential to represent the TSS data to meet the users’ interests. Otherwise, the database becomes no more than a confusing compilation of massive TSS data. First, we masked the clusters with very low expression levels (<5 ppm at the default setting, although there is an option to show all the TSSs), considering they might be derived from intrinsic transcriptional noise of the cells (10) or other experimental errors. Second, we categorized the TSCs in a series of the TSS Seq data so that users can empirically understand the differential usage of the TSSs in different cell types or culture conditions. For graphical representation, we developed a series of new interfaces as shown in Figures 1 and 2. The TSSs corresponding to a particular gene of interest to the user (Figure 1B) can be retrieved and their differential usage in different cellular circumstances can be represented. Figure 1D exemplifies the tissue-specific alternative promoter. In the zinc finger protein 622 gene (NM_033414), the second upstream alternative promoter (moss green) was selectively used in fetal heart, while a different alternative promoter (light green) is used in the other tissues including adult. Also, using our new search page as shown in Figure 1C, users can search promoters showing significant expression changes in response to particular environmental changes.
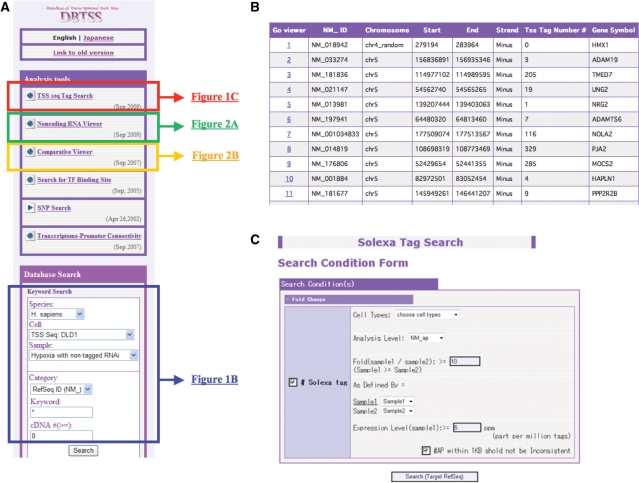
Figure 1.
Interfaces of the newly implemented ‘TSS tag viewer’. TSS sequence tag information can be retrieved from the top page (A) by following either of the links. The viewers corresponding to each link are represented in the indicated figures. (B) In the ‘Database Search’ form, users can directly specify the 5′-end tags of a gene or a cell type they want to view. (C) In the ‘TSS tag search’ form, users can search TSS tags by specifying cell types, fold induction and/or tag counts. They can also choose which category of tags should be considered (e.g. whether tags of different alternative promoters should be counted separately or not). (D) An example of developmental stage-specific alternative promoters. In the zinc finger protein 622 gene (NM_033414), the promoter indicated in moss green (second panel) is selectively used in fetal heart. The upper and lower panels represent the TSS tag usages in adult and fetal tissues, respectively. Height of the vertical bars represents the number of TSS Seq tags located in the corresponding genomic regions. Different alternative promoters are represented by different colors. Each horizontal line represents the experimental condition from which TSS tags were derived. Legends for the tissues and sum of the TSS tag counts are shown at the right margin. (E) Example of the case in which alternative promoter-specific induction was observed in response to IL-4 stimulation in Ramos cells. In the hypothetical protein LOC746 gene (NM_ 014206), the alternative promoter indicated in red (first panel) is selectively induced while the other alternative promoter indicated in blue (second panel) remained unchanged. The indicated TSS regions are magnified to the nucleotide level in the bottom lower panels.
Figure 2.
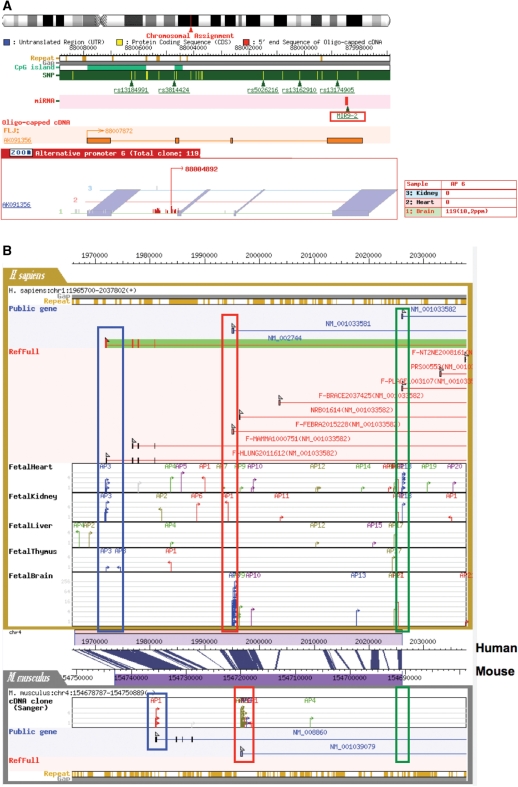
Interface of the updated ‘ncRNA viewer’ and ‘Comparative Genomic viewer’. (A) Example of the TSS tags identified from the surrounding regions of reported small RNAs. The result of the search for a small ncRNA, miR9-2, is shown. Complete cDNA (AK091356) identified in the same region is also represented by orange boxes. (B) Evolutional conservation of the alternative promoters of the protein kinase C zeta (PRKCZ) genes (NM_002744). Different alternative promoters are marked by different colors. Upper and lower panels represent the TSS information in humans and mice, respectively. Corresponding genomic sequences were aligned according to the UCSC Genome Browser information.
Also for example, users can search for all alternative promoters with more than a 5-fold induction after IL-4 stimulation in Ramos cells and with expression level >5 ppm. Figure 1E shows the result of such a search. In the hypothetical protein LOC746 gene (NM_014206), second alternative promoter (red) was selectively induced, while the expression level of the other downstream alternative promoter (blue) remained unchanged. It should be noted that expression analysis using microarrays or RT–PCR could miss such promoter-specific expression changes depending on the positions of the designed DNA probes or PCR primers. To the best of our knowledge, there is no database which represents differential usage of each of the promoters under different experimental conditions in a quantitative manner. Recent studies have suggested such diverse transcriptional regulations give molecular basis to produce complex functional network of human genes by a limited number of total genes (7–9). Also, precise identification of the changes in gene expression associated to each alternative promoter is essential to interpret accumulating ChIP-Seq data (11), for example, to attribute transcription induction to proximal binding of a particular transcription factor. Updated DBTSS will meet the versatile requirements of the analysis of transcriptional network of human genes in the next generation sequencing era.
TSS information for non-coding RNAs
The TSS information collected in an unbiased-manner throughout the human genome is also useful to identify and characterize hitherto unidentified transcripts. Particularly, the new version of DBTSS can be a unique and important resource for identifying primary transcripts of miRNAs and other non-coding RNAs (ncRNAs) which are located in intergenic regions (12). Although hundreds of putative ncRNAs have been identified and their biological characterization undertaken, there have been only few cases in which the TSSs of the primary transcripts the ncRNAs were identified and their promoter structures were elucidated. The new DBTSS contains information of the miRBase database (13) and NR transcript information of the RefSeq database (14), and TSSs within an arbitrary defined distance from the ncRNAs can be retrieved. As shown in Figure 2A, a possible TSS of a primary transcript of an miRBase miRNA miR9-2 (miRBase id=MI0000467; http://microrna.sanger.ac.uk/cgi-bin/sequences/mirna_entry.pl?acc=MI0000467) was found in 6 Kb upstream regions of miR9-2. In addition, our cDNA sequence data also suggested that this miRNA exists in the 3′-end terminal region of a putative non-coding transcript, AK091356.
Updated comparative genomics viewer
Although DBTSS now includes an unprecedented amount of data, we were concerned that many promoters and ncRNAs could be products of ‘transcription noise’, which might occur in the human genome, and thus have no biological relevance. In order to address this concern, we updated our comparative genomic viewer so that the users can examine the evolutional conservation of the surrounding genomic sequences and the TSS tag information against other mammals. Figure 2B exemplifies the case in the protein kinase C zeta (PRKCZ) gene (NM_002744). This gene contains at least three alternative promoters as highlighted in blue, red and green. The most upstream and the third promoters (blue and green) are used in fetal kidney and heart, while the second promoter (red) is selectively used in fetal brain. The genomic sequence of the first two promoters (blue and red) are well-conserved between human and mouse and corresponding TSS tags were observed in both species. In contrast, the genomic sequence surrounding the third promoter (green) is not conserved, and this promoter does not seem to be used in mouse. In order to delineate complex transcriptional regulations of this gene, it is essential to consider different usage and different level of the evolutional conservation of each of the promoters as represented here.
Similarly, we examined evolutionary conservation of the intergenic TSCs of >5ppm and found that at least half are not conserved between human and mouse (detailed analysis will be published elsewhere). It is still not clear whether these intergenic clusters of transcription starts correspond to protein coding or non-coding RNAs performing human-specific biological functions or not. But the information provided in the comparative display should give useful clues to prioritize the targets and design future experiments aiming at further functional studies.
FUTURE PERSPECTIVES
We will continue further updating the next generation sequencing data for more tissues, cells and experimental conditions in humans and mice. We have also started collecting similar data from other model organisms, ranging from various kinds of monocellular eukaryotes, worms, insects, invertebrates and vertebrates as well. On the other hand, in order to clarify the biological relevance of the promoters identified in DBTSS, we started generating RNA Seq (15) data using RNA extracted from nuclear, cytoplasm and translating polysome fractions (16). Such data will reveal which products are actually translated into proteins and in which subcellular compartment the ncRNAs are localized. We believe such integrative transcriptome data will give users the expanded knowledge needed for biological interpretation of each initiation of transcription event.
FUNDING
New Energy and Industrial Technology Development Organization (NEDO) project of the Ministry of Economy, Trade and Industry (METI) of Japan, the Japan Key Technology Center project of METI of Japan, and a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charge: Institute for Bioinformatics Research and Development (BIRD), Japan Science and Technology Agency (JST), Japan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Ms Etsuko Sekimori for excellent programming work. They are also thankful to Dr Alexis Vandenbon for critically reading the manuscript. Computational time was provided by the Super Computer System at the Human Genome Center, Institute of Medical Science, The University of Tokyo.
REFERENCES
- 1.Wakaguri H, Yamashita R, Suzuki Y, Sugano S, Nakai K. DBTSS: database of transcription start sites, progress report 2008. Nucleic Acids Res. 2008;36:D97–D101. doi: 10.1093/nar/gkm901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki Y, Sugano S. Construction of a full-length enriched and a 5′-end enriched cDNA library using the oligo-capping method. Methods Mol. Biol. 2003;221:73–91. doi: 10.1385/1-59259-359-3:73. [DOI] [PubMed] [Google Scholar]
- 3.Tsuchihara K, Suzuki Y, Wakaguri H, Irie T, Tanimoto K, Hashimoto S, Matsushima K, Mizushima-Sugano J, Yamashita R, Nakai K, et al. Massive transcriptional start site analysis of human genes in hypoxia cells. Nucleic Acids Res. 2009;37:2249–2263. doi: 10.1093/nar/gkp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJ, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–177. doi: 10.1016/j.tig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Landry JR, Mager DL, Wilhelm BT. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 2003;19:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willingham AT, Gingeras TR. TUF love for “junk” DNA. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Farnham PJ. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15(Spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 13.Xie J, Zhang M, Zhou T, Hua X, Tang L, Wu W. Sno/scaRNAbase: a curated database for small nucleolar RNAs and cajal body-specific RNAs. Nucleic Acids Res. 2007;35:D183–D187. doi: 10.1093/nar/gkl873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]