Abstract
We have developed phiSITE, database of gene regulation in bacteriophages. To date it contains detailed information about more than 700 experimentally confirmed or predicted regulatory elements (promoters, operators, terminators and attachment sites) from 32 bacteriophages belonging to Siphoviridae, Myoviridae and Podoviridae families. The database is manually curated, the data are collected mainly form scientific papers, cross-referenced with other database resources (EMBL, UniProt, NCBI taxonomy database, NCBI Genome, ICTVdb, PubMed Central) and stored in SQL based database system. The system provides full text search for regulatory elements, graphical visualization of phage genomes and several export options. In addition, visualizations of gene regulatory networks for five phages (Bacillus phage GA-1, Enterobacteria phage lambda, Enterobacteria phage Mu, Enterobacteria phage P2 and Mycoplasma phage P1) have been defined and made available. The phiSITE is accessible at http://www.phisite.org/.
INTRODUCTION
Bacteriophages, though very simple in composition and replication, are the most abundant biological entities on earth. They are the main force in global carbon cycle, in evolution of bacterial species and in maintenance of balance of bacteria in a whole biosphere. The amount and turnover of bacteriophages in the world can be illustrated on the fact, that phage predation destroys an estimated half of the world bacteria population every 48 h (1). Extreme natural adaptability of phages and their strict (or broad) specificity in host bacteria infection make phages ideal adepts for combating human (or other) bacterial diseases. This approach, generally termed as phage therapy, is known to human kind since phage discovery almost a century ago by Twort (2) and d’Herelle (3), but since the advent of chemical antibiotics in the 1940s it has been little used in the West (4).
Bacteriphages were the first organisms studied on a molecular level. In 70-ties, genomes of bacteriophages MS2 and phi-X174 were the first to be completely determined (5,6) and all discoveries of gene regulation are generally based on bacteriophage and bacteria operons research. Over 5500 bacteriophages have been examined in the electron microscope (7). There are 550 completely known phage genomes at the present time. In the EMBL database, entries from ∼1500 different bacteriophages and prophages can be found, giving the approximate number of known and studied bacteriophages. Regulatory elements and gene regulation mechanisms are, however, described only for a few dozens of phage genomes.
Knowing the details about gene regulation is interesting for several reasons. Post-genomic research involves mainly analyzing the dynamics of gene regulation. The commonly accepted assumption that co-regulated genes share similarities in their regulatory mechanism led to a major challenge for the computational biologist—detecting novel regulatory elements (motifs) in such sets of co-expressed genes. These similarities at transcriptional level imply that the promoter region might contain consensus motifs recognized by the same regulatory proteins. In the upstream regions of such sets of co-regulated genes, the common consensus motifs are statistically over-represented as compared to their frequencies in a background set (of non-co-regulated genes) (8). Knowledge of gene regulation systems can lead to several novel practical application ranging from ‘designing of better phages’ used for controlling cellular behavior for medical or biotechnology purposes (9,10) to extremely perspective bio-nanotechnology applications (toggle-switches, oscillators, nano-devices) (9,11,12).
Characterization of gene regulatory networks (GRNs) is quite well summarized for eukaryotes. As an example, we can point out the TRANSFAC (database about eukaryotic transcription factors, their DNA-binding sites and DNA-binding profiles) (13) or The Eukaryotic Promoter Database (14). For prokaryotic organisms there are only few projects under development: PRODORIC (Prokaryotic Database of Gene Regulation) (15) or RegulonDB (transcriptional regulatory network of Escherichia coli K12) (16) covering several hundreds of completely sequenced bacterial genomes. All known information about gene regulation in bacteriophages are spread among scientific papers and books only, partially in primary DNA and protein databases and have not yet been collected in a form of publicly available database. To address this deficiency we have developed phiSITE, database of gene regulation in bacteriophages described in this article.
DATABASE CURRATION AND CONTENT
phiSITE (release 2009.3) contains detailed information about 714 experimentally confirmed or predicted regulatory elements from 32 bacteriophages form Siphoviridae, Myoviridae and Podoviridae families (Table 1). Data related to phage gene regulation are extracted primarily from scientific papers but also from other scientific publications and primary databases. Particular focus is on experimentally confirmed regulatory sites, though predicted sites are also harvested. Many predicted sites in phage genomes are so widely accepted by scientific community that no further experimental evidence is expected. To easily separate entries according to the evidence, experimental/predicted flag of sites is clearly marked in all search results, giving possibility to select and/or analyze only experimental or predicted entries. Phage genome data are parsed from the EMBL database entries using semi-automated parser. All additional data are inserted by curators into the MySQL database back-end using web forms. phiSITE is available to any individual and for any purpose and it is distributed under the ‘Creative Commons Attribution-Share Alike 3.0 Unported License’ (http://creativecommons.org/licenses/by-sa/3.0/).
Table 1.
Statistics of the phiSITE content (Release 2009.3)
| Collected phages (with complete genome) | 32 (29) |
| Myoviridae | 5 |
| Podoviridae | 18 |
| Siphoviridae | 9 |
| Regulatory sites (experimentally identified) | 714 (423) |
| Promoters | 482 |
| Operators | 61 |
| Terminators | 165 |
| Attachment sites | 6 |
| Source publications | 127 |
The base element of phiSITE is defined as a site, representing one regulatory element present on a phage genome. This can be either promoter, operator, transcription terminator or attachment site. Site element can be segmented into several subsites (if known), particular cis-regulatory signals (e.g. −35 and −10 for prokaryotic promoter). The database also provides references to the method of evidence for experimentally confirmed sites. All sites are linked to the other phiSITE tables describing the phage and its features. Information about complete phage genome is also included (if available), together with names and positions of all known genes. phiSITE keeps also updated information about phage and phage host taxonomy, together with numerous links to other database resources described in section ‘Phage genome browser’ below. There are also several accompanying analyzing tools under development, accessible in the Tools section. These include:
PSSM-convert: a tool for creation and conversion of Position Specific Scoring Matrices in different formats.
Free Energy: a tool for computation of Gibbs free energy distribution in DNA sequence.
Promoter Hunter: a tool for promoter search in prokaryotic genomes.
Each tool is accompanied with corresponding help instructions, and their detailed description is beyond the scope of this paper.
The phiSITE database is permanently updated and new releases are published several times a year.
DATABASE ACCESS
The main access to the database is provided via the web interface at http://www.phisite.org/. The phiSITE portal is based on a well-established LAMP platform (Linux/Apache/MySQL/PHP). Users can utilize several ways to approach the data:
searching and exporting the entries via Quick Search and Advanced Search;
exploring phage genomes via graphical applet in the Phages section;
exploring phage GRNs via BioTapestry Viewer;
browsing and exporting the entries according to the phage or host taxonomy in the Browse section; and
downloading the whole content of the database in XML format in Downloads section.
Searching the entries
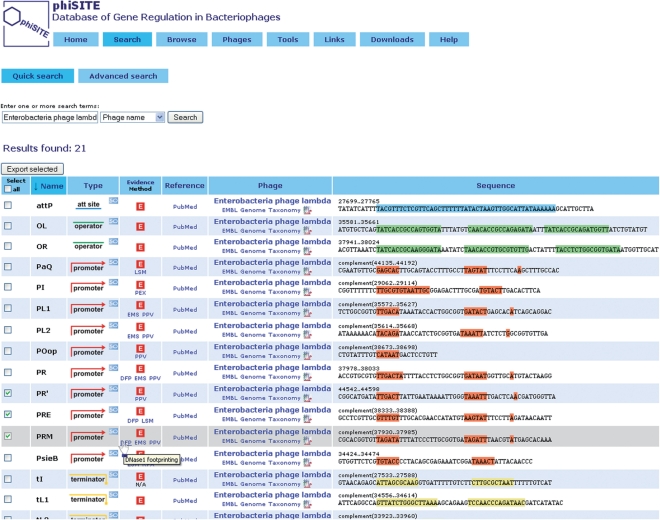
User can search the content of a database using ‘Quick Search or Advanced Search’. Search terms are looked up either in all text fields (phage name, host name, site name, site description or site type) or in a single field selected by a user. In ‘Advance Search’ different search fields for each search term can be specified, with an optional usage of wildcards. Search results are provided in a form of table with customizable order. Each entry includes site name, type (promoter, operator, terminator or attachment site), method of evidence, source reference, phage details and semi-graphical representation of DNA segment containing the site (Figure 1). All sites are linked to the Sequence Ontology thesaurus (17). Arbitrary number of entries from search result page can be manually selected and exported using exporting module described in the section ‘Browsing and exporting the entries’ below.
Figure 1.
Result of a ‘Quick Search’ for all regulatory sites from Enterobacteria phage lambda. Each entry contains description of a site and links to other information resources. Three sites (PR', PFE and PRM) are selected and can be exported using the ‘Export selected’ button.
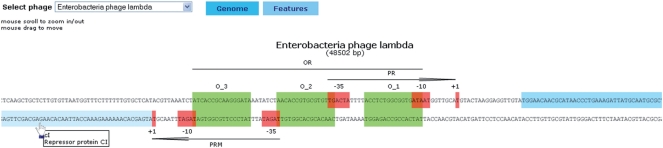
Phage genome browser
The system possess proprietary graphical genome browser (Figure 2). It is used to visualize all phages with known and annotated genome. It is based on Adobe Flash technology (http://www.adobe.com/products/flash/) and it is dynamically linked to the phiSITE MySQL back-end. Genome browser provides a graphical representation of all phage genes and regulatory sites where all elements are zoomable up to the primary sequence level. User can use a mouse to zoom in/out and to drag along the genome sequence. All elements are labeled with a name and a short description. Features section contains phage and phage host taxonomic classification, provides set of links to related bioinformatics resources (EMBL, UniProt, NCBI taxonomy database, NCBI Genome, ICTVdb and PubMed Central) (18–21) and also to other sections of phiSITE portal: BioTapestry viewer (for selected phages) and direct link to the list of all sites associated with a particular phage.
Figure 2.
Graphical applet zoomed to the region between Enterobacteria phage lambda genes cI and cro. User can use a mouse to zoom in/out and to scroll along the phage genome.
BioTapestry viewer
We have adapted BioTapestry tool for visual representation of phage GRNs. BioTapestry is a free and open source Java based interactive tool for building, visualizing and simulating GRNs (22). It can output regulatory network in SBML format (23), which can be read into a GRNs simulation environment such as Dizzy (24). Source data for visualization in BioTapestry Editor are imported as Comma Separated Value files from phiSITE back-end, where interaction instructions extracted from scientific literature are defined. Source type ‘gene’ is used for genes and gene products, and source type ‘box’ for regulatory sites. Several types of interactions are described in the BioTapestry model: (i) initiation of transcription of a gene from promoter, (ii) activation of transcription by a product of phage gene, (iii) repression of transcription by a product of a gene binding to the operator of target promoter, (iv) repression of transcription by the operator negatively influencing promoter, (v) termination of transcription initiated from the promoter and (vi) antitermination of transcription by a product of antiterminator gene. Positive regulation is depicted as an arrowed line pointing from the master to the slave element (i–iii), negative regulations as a ‘T’ shaped line pointing to the slave element (iv,vi) and neutral relation as a straight line between master and slave elements (v). The Editor automatically creates a network of interactions and assembled model is made available on the web using Java Web Start technology. Only interactions among the phage genome elements are defined at the moment, though future versions may also include phage host regulatory elements. Example of Enterobacteria phage lambda regulatory region is given in Supplementary Data.
Browsing and exporting the entries
Set of phiSITE entries can be exported using dynamic export module and used in further analyses in a variety of bioinformatics tools. User can select a group of sites according to the phage or phage host taxonomic hierarchy. Evidence (experimental, predicted or both) and site and subsite types can also be selected. Each taxonomic selection step is coupled with background counting of sites currently selected. After selection, user has an option (i) to build a motif representation for selected sites, (ii) to export sites as FASTA sequences or (iii) to export selected site in XML format. Selecting Build motif representation is followed by a sequence alignment assembly process mediated by a ClustalW2 algorithm (25) and the motif is exported in several output formats: TRANSFAC database (13), FASTA, Patser (26), PromScan (27), Postion Weight Matrix (26) and Sequence logo (28). XML format is based on XML version 1.0 specification and the output file is coupled with XML Document Type Definition (DTD).
CONCLUSION
phiSITE is a manually curated database dedicated to the gene regulation in bacteriophages. It is the first resource of this kind and it is freely available to all potential users. Mainly experimentally detected cis-regulatory elements on phage genomes are harvested from scientific articles. This data are accompanied with additional information about phages and phage hosts, external links and associated tools. Curation and update process of phiSITE database will be continued. Further enhancements will include improved visualization models for selected bacteriophages with possible application in systems biology simulation engines, implementation of web services to access the data. Next version of genome browser will also cover direct link to the description of genes and regulatory elements, mediated by clicking the corresponding element in the browser and also improved graphical rendering of visualized entities. We are awaiting response from scientific community in order to improve the services provided by the phiSITE platform.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Slovak Research and Development Agency [grant number APVT-51-025004]; Scientific Grant Agency of the Ministry of Education of the Slovak Republic and of Slovak Academy of Sciences [grant number VEGA 2/0100/09].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank their database curators Ida Baumgartnerova, David Porubsky and Diana Hierwegova for literature mining and keeping the data up to date. The authors are grateful to Petra Polovkova for assistance in MySQL database design and implementation. Finally, the authors would like to thank Renata Novakova for sharing her scientific experiences in the field of Gene Expression.
REFERENCES
- 1.Hendrix RW. Bacteriophages: evolution of the majority. Theor. Popul. Biol. 2002;61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- 2.Twort FW. An investigation on the nature of ultra-microscopic viruses. Lancet. 1915;186:1241–1243. doi: 10.1017/s0022172400043606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.d’Herelle F. Sur un microbe invisible antagoniste des bacilles dysenteriques. CR Acad. Sci. Paris. 1917;165:373–375. [Google Scholar]
- 4.Housby JN, Mann NH. Phage therapy. Drug Discov. Today. 2009;14:536–540. doi: 10.1016/j.drudis.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, Min Jou W, Molemans F, Raeymaekers A, Van den Berghe A, et al. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260:500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- 6.Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 7.Ackermann HW. 5500 bacteriophages examined in the electron microscope. Arch. Vrol. 2006;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 8.Thijs G, Lescot M, Marchal K, Rombauts S, De Moor B, Rouzé P, Moreau Y. A higher-order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics. 2001;17:1113–1122. doi: 10.1093/bioinformatics/17.12.1113. [DOI] [PubMed] [Google Scholar]
- 9.Hasty J, McMillen D, Isaacs F, Collins JJ. Computational studies of gene regulatory networks: in numero molecular biology. Nature Rev. Genetics. 2001;2:268–279. doi: 10.1038/35066056. [DOI] [PubMed] [Google Scholar]
- 10.Skiena SS. Designing better phages. Bioinformatics. 2001;17:S253–S261. doi: 10.1093/bioinformatics/17.suppl_1.s253. [DOI] [PubMed] [Google Scholar]
- 11.Shu D, Huang LP, Hoeprich S, Guo P. Construction of phi29 DNA-packaging RNA monomers, dimers, and trimers with variable sizes and shapes as potential parts for nanodevices. J. Nanosci. Nanotechnol. 2003;3:295–302. doi: 10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- 12.Taton TA. Bio-Nanotechnology: two-way traffic. Nature Materials. 2003;2:73–74. doi: 10.1038/nmat824. [DOI] [PubMed] [Google Scholar]
- 13.Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief. Bioinform. 2008;9:326–332. doi: 10.1093/bib/bbn016. [DOI] [PubMed] [Google Scholar]
- 14.Schmid CD, Perier R, Praz V, Bucher P. EPD in its twentieth year: towards complete promoter coverage of selected model organisms. Nucleic Acids Res. 2006;34:D82–D85. doi: 10.1093/nar/gkj146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grote A, Klein J, Retter I, Haddad I, Behling S, Bunk B, Biegler I, Yarmolinetz S, Jahn D, Munch R. PRODORIC (release 2009): a database and tool platform for the analysis of gene regulation in prokaryotes. Nucleic Acids Res. 2008;37:D61–D65. doi: 10.1093/nar/gkn837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gama-Castro S, Jiménez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Peñaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muñiz-Rascado L, Martínez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2007;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cochrane G, Akhtar R, Bonfield J, Bower L, Demiralp F, Faruque N, Gibson R, Hoad G, Hubbard T, Hunter C, et al. Petabyte-scale innovations at the European Nucleotide Archive. Nucleic Acids Res. 2009;37:D19–D25. doi: 10.1093/nar/gkn765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Büchen-Osmond C. Manual of Clinical Microbiology. 8th edn. Vol. 2. Washington DC: ASM Press; 2003. Taxonomy and classification of viruses; pp. 1217–1226. [Google Scholar]
- 22.Longabaugh W JR, Davidson EH, Bolouri H. Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Develop. Biol. 2009;283:1–16. doi: 10.1016/j.bbagrm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 24.Ramsey S, Orrell D, Bolouri H. Dizzy: stochastic simulation of large-scale genetic regulatory networks. J. Bioinform. Comput. Biol. 2005;3:415–436. doi: 10.1142/s0219720005001132. [DOI] [PubMed] [Google Scholar]
- 25.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 26.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 27.Studholme DJ, Dixon R. Domain architectures of sigma54-dependent transcriptional activators. J. Bacteriol. 2003;185:1757–1767. doi: 10.1128/JB.185.6.1757-1767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]