Abstract
The MiST2 database (http://mistdb.com) identifies and catalogs the repertoire of signal transduction proteins in microbial genomes. Signal transduction systems regulate the majority of cellular activities including the metabolism, development, host-recognition, biofilm production, virulence, and antibiotic resistance of human pathogens. Thus, knowledge of the proteins and interactions that comprise these communication networks is an essential component to furthering biomedical discovery. These are identified by searching protein sequences for specific domain profiles that implicate a protein in signal transduction. Compared to the previous version of the database, MiST2 contains a host of new features and improvements including the following: draft genomes; extracytoplasmic function (ECF) sigma factor protein identification; enhanced classification of signaling proteins; novel, high-quality domain models for identifying histidine kinases and response regulators; neighboring two-component genes; gene cart; better search capabilities; enhanced taxonomy browser; advanced genome browser; and a modern, biologist-friendly web interface. MiST2 currently contains 966 complete and 157 draft bacterial and archaeal genomes, which collectively contain more than 245 000 signal transduction proteins. The majority (66%) of these are one-component systems, followed by two-component proteins (26%), chemotaxis (6%), and finally ECF factors (2%).
INTRODUCTION
In all living organisms, signal transduction systems link extracellular signals to appropriate cellular responses. In multicellular eukaryotes, hormones, cyto- and chemokines, and neurotransmitters comprise the major signaling molecules. In prokaryotes and unicellular eukaryotes, the major signals consist of various physico-chemical parameters of the environment: small molecules, light, oxygen, temperature, and other stimuli. The molecular machinery that transmits intracellular signals is quite different in eukaryotes and prokaryotes. Eukaryotes utilize rather complex signal transduction cascades that are usually initiated by G-protein coupled receptors and ion channels, while prokaryotes employ simpler signal transduction systems that typically consist of one or two proteins (1,2). Historically, the best studied mode of prokaryotic signal transduction is two-component signaling (1). In a typical two-component system, the input domain of the sensor histidine kinase detects an environmental signal, which results in the activation of the kinase domain followed by autophosphoryation at a specific histidine residue. The phosphoryl group is then transferred to a specific aspartate residue in the receiver domain of the second protein, a response regulator (RR). This phosphorylation event activates the output domain of the RR, which triggers the cellular response. Most output domains in bacterial RR bind DNA and therefore act as transcriptional regulators (3); however, many other output domains have enzymatic activities, for example, regulating the concentration of the second messenger, cyclic di-GMP (4). Some RR do not have a distinct output domain and their regulatory function resides within the receiver domain. A classic example of such a response regulator is a chemotaxis protein, CheY, which controls the rotational direction of the bacterial flagellar motor (5).
A comprehensive genomic study of more than a hundred microbial genomes revealed that two-component regulatory systems are not the prevalent mode of signal transduction in prokaryotes (3). The majority of signal transduction systems in prokaryotes consist of a single protein that contains both input and output domains, but lacks phosphotransfer domains typical of two-component signaling. Such regulatory proteins were termed “one-component systems” (3). On average, microbial genomes contain many more one-component than two-component systems. In addition to these two principal modes of signal transduction in prokaryotes, other signaling elements are recognized. The chemotaxis signal transduction system (5) involves several specialized proteins in addition to a specialized version of a histidine kinase. Therefore, it is often viewed as a unique case of two-component signaling. Finally, recent studies implicated extracytoplasmic function (ECF) sigma factors as yet another mode of signal transduction in prokaryotes (6–8).
Due to its functional diversity (linking numerous signals to various types of responses via different modes) that translates into extreme sequence variation of input and output domains, pronounced domain shuffling, and variability of interacting modules, signal transduction presents a major challenge for genomic identification and logical classification. The only automated procedure for identifying signal transduction proteins is implemented in the Clusters of Orthologous Groups of Proteins (COG) database (9) as a special “Signal Transduction” category; however, in contrast to other categories of this resource, the “Signal Transduction” category contains many erroneous annotations, due to the reasons outlined above. Because COG is an integral part of many genome annotation platforms, these flawed annotations propagate to downstream analyses.
To provide a better classification system and an electronic resource that can be used for genome annotation, we have designed MiST, a Microbial Signal Transduction database (10). Since its launch in 2006, MiST has been accessed by more than 5000 unique users and was used for annotation in several genome projects (11–17). These results establish MiST as a useful community database and genome annotation tool. Our continuous efforts in improving MiST resulted in the new database release (MiST2 presented here) with many new features and significant improvements.
NEW FEATURES AND IMPROVEMENTS
Draft genomes
The original MiST database only supports the analysis of completely sequenced genomes; however, many genome sequencing projects result in intermediate assemblies. Despite incomplete coverage and some inaccuracies due to the preliminary nature of this data, many researchers are interested in the signal transduction properties of such draft genomes. Furthermore, some incomplete genomes are never closed/finished and provide the only genome record available. Thus, we have enhanced our database implementation and annotation pipeline to support the analysis of draft genomes. Available draft genomes with Refseq annotations are downloaded from NCBI and processed in a similar fashion as complete genomes; however, instead of complete chromosomes and plasmids, draft genomes typically consist of tens to hundreds of individual contigs. Distinct draft versions of the same genome are handled as separate genomes, which enables the comparison of different genome versions.
Extra-cytoplasmic function (ECF) proteins
A specialized subunit of the bacterial holoenzyme, sigma (σ) factors direct RNA polymerase to bind to specific promoter sequences. In addition to a primary, housekeeping σ factor, bacteria may also contain alternative σ factors that substitute for the core σ factor and redirect transcription to alternative promoter binding sites (18). ECF σ factors, the smallest and most diverse sub-family of σ70 proteins, consist of only two domains, σ2 and σ4, which participate both in RNA polymerase interaction and recognition of the bipartite sequence motif of the alternative promoter site. ECF σ factors comprise the largest group of alternative σ factors and represent the third most abundant bacterial signaling mechanism. In the absence of a suitable stimulus, ECF σ factors are usually bound by a cognate anti-σ factor and thereby kept inactive. Upon receiving an appropriate environmental signal, the anti-σ factor releases the ECF σ factor, which then redirects transcription after binding to the RNA polymerase (6,7).
Staroń et al. performed a comprehensive phylogenomic analysis of the ECF σ factor protein family and devised a profile-based system for identifying and classifying these unique signaling proteins into distinct ECF groups (8). Using their collection of 44 group-specific hidden Markov models (HMMs) and classification system, we identify and catalog ECF σ factors for each genome in addition to one- and two-component signaling proteins. The full annotation of each ECF group is included in MiST2 and linked from annotated ECF σ factors.
Updated signaling profile collection
All signal transduction proteins are identified based on the presence and/or absence of specific signaling domain profiles. In addition to the ECF HMMs, we have expanded the collection of signaling profiles to 166 Pfam domains (19) and created a custom signaling library, Agfam (Supplementary Table S1). The Agfam library currently consists of a receiver domain profile (RR) that models the conserved receiver domain of response regulators and 24 profiles that represent the catalytic (transmitter) domain (HK_CA) of histidine kinases (HK). Our approach for generating these profiles relies on exhaustive PSI-BLAST searches seeded with bona-fide family members (those with known 3D structures) to identify homologous sequences and careful semi-automatic construction of multiple sequence alignments (Figure 1). Due to the extreme conservation of receiver domains, we were able to produce a quality alignment of all 26 492 receiver domain sequences found in the UniProt database (19 May 2008). In contrast, the transmitter domain of HK is significantly less conserved and it was not possible to produce a single high-quality alignment of all HK_CA domains. Instead, we generated 23 individual profiles (labeled HK_CA:1–23, covering 16 829 sequences) that correspond to specific clusters of closely related members, and a general profile, which represents the overall sequence characteristics of the histidine kinase catalytic domain family.
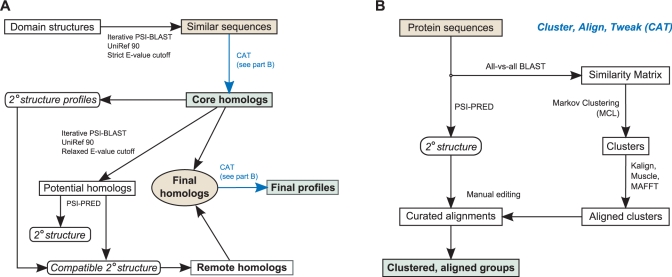
Figure 1.
Semi-automatic algorithm for defining high-quality domain models. (A) Bona fide domain members which have had their structure solved are subjected to iterative PSI-BLAST searches (30) against the UniRef90 (31) database with a stringent E-value threshold. The resulting sequences are then clustered, aligned and edited (CAT, part B) to form the set of core homologs. Remote homologs are identified by the same procedure with a much relaxed threshold and then removing hits that do not match a secondary structure type associated with at least one core homolog. The resulting remote homologs are combined with the core homologs and then subjected to the CAT process to produce the final domain model(s). (B) The CAT sub-algorithm is a divide-and-conquer method for addressing the extreme sequence divergence present in signal transduction families. Markov Clustering Linkage (32) simulates a random-walk through all-versus-all BLAST results and produces clusters of related members. After aligning and editing each individual subgroup, they are further combined into one or more final curated alignments.
We evaluated the performance of our HK_CA profiles versus the Pfam HATPase_c domain (accession PF02518) using a test set of 875 complete bacterial and archaeal genomes (July 2009). Overall, the HK_CA profiles demonstrated similar sensitivity to HATPase_c, yet superior specificity for sensor histidine kinase proteins. The seed alignment for the HATPase_c domain contains 662 sequences extracted from the SwissProt database and models the ATPase domains of HK, DNA gryases, heat-shock proteins, topoisomerases, phytochrome-like ATPases, and DNA mismatch repair proteins. Consequently, the HATPase_c profile is only 90% specific for sensor kinase proteins as opposed to 99.8% for our HK_CA profiles. We attribute the higher specificity of HK_CA to two factors. First, the seed alignments do not contain any functionally unrelated (e.g. gyrase, heat-shock proteins, etc.) sequences. Second, the HK_CA seed alignments consist of nearly 17 000 sequences drawn from a much larger protein space and thus are much more representative. A HMMER search (20) with the RR profile against the proteomes associated with 875 complete genomes revealed 32 162 instances of this domain. In contrast, HMMER searches with the Pfam Response_reg model identified only 31 149 receiver domains. Only 56 of the Response_reg hits were not found by our RR profile. Conversely, 1070 receiver domains were uniquely recognized by the RR profile. These results substantiate that our RR profile is the most sensitive means for identifying RR.
Improved classification
The original MiST database (10) simply grouped signal transduction proteins into two categories, one- and two-component proteins, and provided a basic interface for browsing these proteins. Using the expanded signaling domain repertoire described earlier, we have significantly enhanced our domain-based classification with a hierarchical rule system for more specifically classifying these major modes of signal transduction (Table 1). We identify proteins comprising two-component regulatory systems by first searching for matches to the conserved transmitter and receiver domains of HK and RR, respectively. Based on the presence and absence of these hallmark domains, detected two-component proteins are further classified into the following five distinct groups: HK, RR, hybrid histidine kinase (HHK), hybrid response regulator (HRR), and “other”. Hybrid two-component proteins contain both transmitter and receiver domains. Relatively few of these proteins have been experimentally studied and no rigorous experimental basis is available for classifying these proteins. Thus, we group them based on the linear position of the transmitter and receiver domains within the protein sequence. HHKs have transmitter domains N-terminally to receiver domains, whereas HRRs have receiver domains N-terminally to transmitter domains. HKs contain a transmitter domain (represented by multiple Pfam and Agfam domain profiles in our list of signaling domains, Supplementary Table S1) and do not contain receiver domains. In contrast, RRs contain a receiver domain but no transmitter domain. Remaining proteins that are recognized as parts of two-component signaling (e.g. stand-alone histidine phosphotransfer domains recognized by the Hpt profile, accession PF01627) are placed in the “other” category.
Table 1.
Domain-based rules for classifying signal transduction proteins
| Rule | Classification |
|---|---|
| Pfam or Agfam marker domain | |
| Chemotaxis domains | |
| HK_CA:Che (Agfam) | Chemotaxis, CheA |
| CheW and transmitter domain | Chemotaxis, CheA |
| CheW and receiver domain | Chemotaxis, CheV |
| CheW | Chemotaxis, CheW |
| CheB_methylest | Chemotaxis, CheB |
| CheR or CheR_N | Chemotaxis, CheR |
| CheD | Chemotaxis, CheD |
| CheZ | Chemotaxis, CheZ |
| CheC and not SpoA | Chemotaxis, CheCX |
| MCPsignal | Chemotaxis, MCP |
| * | Chemotaxis, Other |
| Transmitter or receiver domain | |
| HATPase_c signaling domain + receiver | |
| HATPase_c before receiver | Two-component, HHK |
| N-terminal receiver | Two-component, HRR |
| * | Two-component, other |
| HATPase_c signaling domain | Two-component, HK |
| Receiver domain | Two-component, RR |
| * | Two-component, other |
| Output domain | One-component |
| * | Other |
| ECF domain | ECF |
The rule system is hierarchical and each rule is processed sequentially. Proteins are classified according to the first matching rule. Asterisks (*) match all proteins. The complete list of Pfam and Agfam signaling domains is provided in Supplementary Table 1. Marker domains represent signaling domains that implicate a protein as participating in signal transduction. Unless otherwise indicated, all domains are from Pfam (19). HHK – hybrid histidine kinase, HRR – hybrid response regulator, HK – histidine kinase, RR – Response regulator.
Two special cases exist for filtering out non-signaling proteins. First, in addition to the catalytic domain of HK, the Pfam HATPase_c seed alignments contain sequences belonging from structurally related ATPase proteins (see above) that do not participate in signal transduction. Consequently, searches with the HATPase_c profile rank these proteins as statistically significant. We filter out these proteins which contain a HATPase_c signaling domain in combination with any of the following Pfam models: DNA_gyraseA_C, DNA_gyraseB, DNA_gyraseB_C, Toprim, HSP90, DNA_topoisoIV, DNA_mis_repair, MutL_C, and Topo-VIb_trans. Second, the hydrolase domain, HD, which is implicated in signal transduction (21), frequently occurs in other non-signaling proteins. We exclude HD-containing proteins from the list of signal transduction proteins if they also contain the following non-signaling Pfam domains: PolyA_pol, TGS, RelA_SpoT, KH_1, KH_2, tRNA_anti, PPx-GppA, DEAD, Helicase_C, and tRNA-synt_1d.
Chemotaxis is the best studied prokaryotic signaling pathway, which typically consists of several key proteins that functionally link an environmental signal with motility (5). Because the chemotaxis pathway is based on a specialized version of a histidine kinase (CheA) and a response regulator (CheY), it is considered a distinct part of two-component signaling (2). Based on the work performed by Wuichet and colleagues (22), we use specific chemotaxis domain models to classify chemotaxis proteins into the major protein groups: CheA, CheV, CheW, CheR, CheB, CheCX, CheD, CheZ, and “other” (Table 1). Unfortunately, the Pfam profiles, CheZ and CheC, are not well defined and fail to accurately identify all target proteins. Moreover, the CheC profile does not well discriminate between CheC and CheX proteins, and thus proteins with this domain are labeled CheCX (22). Despite these shortcomings, we were able to document and organize more than 15 400 chemotaxis proteins and provide more specific information on this special class of two-component signaling.
Identification and classification of neighboring two-component proteins
Many experimentally studied two-component systems are encoded as pairs on a chromosome, and response regulator genes commonly co-occur adjacent to their regulatory targets (2). Moreover, MiST database searches across all genomes reveal that two-component genes frequently occur (62%) as genomic neighbors (10). Consequently, neighboring two-component genes likely encode functionally coupled two-component systems and are excellent candidates for future experimental research.
Based on these principles, we systematically identify all sets of neighboring two-component genes for each genome and classify them into four basic groups according to their genomic context. All members of a two-component set must belong to the same gene string, which we operationally define as a contiguous set of genes oriented in the same direction (i.e. strand), each separated by no more than 200 base pairs, and not interrupted by any gene encoded in the opposite direction. Within the context of a given gene string, distinct two-component sets must be at least three genes apart and are classified into the following categories: (1) classical—single HK and RR gene pair with 0, 1, or 2 intervening genes; (2) hybrid—identical to classical except a hybrid HK or hybrid RR (see classification rules above) is paired with the RR and HK, respectively; (3) complex—two or more two-component genes with no more than 0, 1, or 2 intervening genes; and (4) orphan—a single two-component gene that is genomically isolated from any other two-component genes.
While not based on a rigorous study of intergenic distances, which are known to vary between species (23), the 200 base pair threshold is intended as a simple, conservative cutoff for establishing gene clusters and is similar to those used in recent studies (24,25). Because gene rearrangements and reshuffling frequently occur between species (26), our approach permits neighboring two-component genes in the same set to be separated by up to two intervening genes.
Advanced protein/gene data retrieval
We have substantially improved the web page for viewing and retrieving information about specific proteins and/or genes (Figure 2). All proteins are visualized using their domain architecture, and it is now possible to retrieve the subsequence of specific domains by simply clicking on the box surrounding a specific domain. The genomic context of the current gene/protein being displayed is visualized in the “Genome neighborhood” section and it is possible to dynamically browse the chromosome/plasmid (complete genomes) or contig (draft genomes) using the mouse. Clicking on neighboring genes transparently updates the annotation details and domain architecture visualization. It is also possible to add or remove the currently viewed gene from the gene cart without leaving the page. We have added the capability to retrieve upstream or downstream DNA of any gene. MiST2 contains indices of external database identifiers to the NCBI non-redundant protein database, Protein Data Bank (27), and UniProt/SwissProt (28) database enabling access to protein structures and additional annotations. These facilitate finding proteins of interest via custom searches and the complete list of cross-references is displayed when viewing a specific protein/gene.
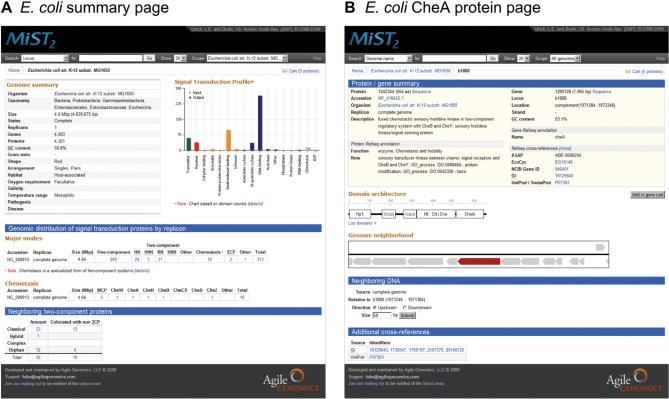
Figure 2.
Screenshots of the MiST2 website. (A) E. coli genome summary page. Below the header and navigational links there are three sections: genome and organism metadata, and a hyperlinked graphical image of the genome’s signal transduction profile; fully linked tables displaying the genomic distribution of one-component, two-component, chemotaxis and ECF signaling proteins by replicon; and lastly a table containing the counts of neighboring two-component proteins. (B) E. coli CheA protein page. The Refseq annotation and database cross-references for the currently viewed protein and corresponding gene is displayed at the top. This is followed by an interactive visualization of the protein’s domain architecture. The genome neighborhood section contains an AJAX-driven, dynamic representation of the genomic context surrounding the currently viewed protein. In the neighboring DNA section, it is possible to retrieve upstream or downstream DNA sequence data. Hyperlinked cross-references to external databases appear at the bottom of the page.
Improved taxonomy browser and genome filters
We have completely overhauled the taxonomy browser and selection tool. All genomes in MiST2 are hierarchically displayed according to their taxonomy and organized with a user-specified taxonomy level. With the tree it is possible to define multiple genome filters that enable searching against specific user-defined groups of genomes. Changing the taxonomy level uses remote AJAX calls to dynamically update the taxonomy tree. In addition to five basic taxonomy levels—kingdom, phyla, class, order, and family—we also provide a “Major taxonomic group” level to facilitate viewing/selecting species at the phyla level and all sublevels of the proteobacterial group.
Miscellaneous new features
MiST2 now supports searching by genome name, taxonomy, MiST2 protein identifier, locus tag, Refseq gene name, Refseq description, Pfam and/or Agfam domain architecture, and several classes of external database identifiers (accession number, GI, SwissProt, UniProt, and PDB). Users may incorporate Boolean full-text logic to form more complex queries when searching against the Refseq description and domain architectures. By default queries are performed against all available genomes, yet it is possible to narrow searches to specific genomes of interest by specifying a user-defined genome filter (created using the taxonomy browser). Available genome filters are displayed in the scope drop-down box of the search panel (Figure 2). The scope drop-down box on the summary page for a specific genome automatically displays a genome filter for the currently viewed genome for restricting searches to this organism. Other improvements include the addition of organism metadata (e.g. habitat, gram stain, etc.), a gene/protein cart for retrieving gene and protein sequences, ranking genomes by the number of hits returned by the Google Scholar service (http://scholar.google.com), a mailing list, and a completely redesigned, biologist-friendly website. Every page includes the search panel for rapidly finding relevant information and breadcrumb navigation for keeping track of one’s location.
DATABASE RESULTS
As of October 2009, MiST2 contains 245 521 signal transduction proteins (Table 2). It has been previously noted that one-component systems dominate prokaryotic signal transduction (3) and our results based on the counts from 1123 distinct genomes support this observation. Indeed, if we estimate the number of two-component systems based on the number of RR, one-component systems outnumber two-component systems by a factor of five. RRs slightly exceed the number of HKs (1.2:1), yet this ratio approximates an even distribution when compared to the combined number of HK and HHK. The complex two-component proteins, HHK and HRR, are far less abundant than HK and RR, respectively, suggesting that most bacteria utilize simpler signaling pathways. The nearly 1:1 ratio of the HK to RR occurrence strongly reinforces the notion that two-component regulatory systems are not branched and typically link a specific signal (or a set of signals in case of multiple input domains) to a specific cellular response. Archaea in general have lower counts of signal transduction proteins than Bacteria. Phylogenetic (29) and genomic (3) analyses suggest that two-component systems have been laterally transferred to Archaea from Bacteria. Therefore, the low counts of two-component proteins in Archaea are not surprising; however, peculiar observation is that Archaea contain significantly fewer HHK than expected (when normalized against the total number of two-component proteins) and significantly more HRR (Table 2). This paradox warrants a special consideration and experimental validation of the role of these hybrid proteins in the biology of Archaea.
Table 2.
Distribution of signal transduction proteins within complete and draft genomes belonging to Archaeal and Bacterial phyla
| Genomes | One-component | Two-component |
ECF | |||||
|---|---|---|---|---|---|---|---|---|
| HK | HHK | RR | HRR | Chemotaxis | ||||
| Archaea | ||||||||
| Complete | 67 | 3265 | 546 | 8 | 304 | 142 | 453 | – |
| Draft | 2 | 77 | – | – | 1 | – | 10 | – |
| Bacteria | ||||||||
| Complete | 899 | 135 396 | 20 862 | 4717 | 26 962 | 923 | 13 549 | 5332 |
| Draft | 155 | 22 217 | 3791 | 364 | 3981 | 61 | 1418 | 784 |
| Total | 1123 | 160 955 | 25 199 | 5089 | 31 248 | 1126 | 15 430 | 6116 |
HK, histidine kinase; HHK, hybrid histidine kinase; RR, response regulator; HRR, hybrid response regulator.
We classified 38 984 genomically neighboring two-component proteins into the following categories: 31 040 (49%) classical, 1872 hybrid (3%), and 6072 (10%) complex sets. The remainder did not neighbor any other two-component protein. The majority (66%) of two-component sets co-occurs with non-signaling proteins and putatively regulates the expression of these neighboring genes or interact with their protein products.
CONCLUSIONS AND FUTURE PLANS
Because of their primary role in nearly all cellular processes, knowledge of signal transduction proteins and pathways will continue to be a driving force in furthering biological discovery. MiST2 is the premier database on microbial signal transduction. This second generation database provides a comprehensive and well-defined system for extensively classifying and documenting the major modes of signal transduction—one-component, two-component, chemotaxis, and ECF—in both complete and draft genomes. The modernized web interface of MiST2 enables researchers to rapidly and conveniently access relevant genomic data. We expect MiST2 to continue building on its previous success as a valuable tool for supporting microbiological research, genome annotation, and applications for biomedical, environmental, and bioenergy research. We are currently analyzing the signal transduction properties of metagenomic data sets and will integrate these results into MiST2. Other developments underway include building an orthologous network of signaling domains, operon and signaling pathway reconstruction, literature-based curation, and continued website development.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM083177 to L.E.U. and GM72285 to I.B.Z., partial); South Carolina Research Association (2008-009 to L.E.U.); BioEnergy Science Center which is supported by the Office of Biological and Environmental Research in the DOE Office of Science. Funding for open access charge: BioEnergy Science Center which is supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Conflict of interest statement. None declared.
ACKNOWLEDGMENTS
We would like to thank the BLAST/STIM community for their support and insight into developing MiST. We are especially grateful to Jan Lukens and “Friends of the Zhulin lab” for financially supporting the purchase of our Linux cluster.
REFERENCES
- 1.Kofoid EC, Parkinson JS. Transmitter and receiver modules in bacterial signaling proteins. Proc. Natl Acad. Sci. USA. 1988;85:4981–4985. doi: 10.1073/pnas.85.14.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 5.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 6.Butcher BG, Mascher T, Helmann JD. Environmental sensing and the role of extracytoplasmic function sigma factors. In: El-Sharoud W, editor. Bacterial Physiology. Berlin, Heidelberg: Springer; 2008. pp. 233–261. [Google Scholar]
- 7.Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 8.Staroń A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol., 10.1111/j.1365-2958.2009.06870.x. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19737356. [DOI] [PubMed] [Google Scholar]
- 9.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulrich LE, Zhulin IB. MiST: a microbial signal transduction database. Nucleic Acids Res. 2006;35:D386–D390. doi: 10.1093/nar/gkl932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chain PSG, Denef VJ, Konstantinidis KT, Vergez LM, Agulló L, Reyes VL, Hauser L, Córdova M, Gómez L, González M, et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl Acad. Sci. USA. 2006;103:15280–15287. doi: 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S, Binder BJ, Edmonds J, Ye W, Orcutt B, et al. Ecological genomics of marine Roseobacters. Appl. Environ. Microbiol. 2007;73:4559–4569. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson I, Rodriguez J, Susanti D, Porat I, Reich C, Ulrich LE, Elkins JG, Mavromatis K, Lykidis A, Kim E, et al. Genome sequence of Thermofilum pendens reveals an exceptional loss of biosynthetic pathways without genome reduction. J. Bacteriol. 2008;190:2957–2965. doi: 10.1128/JB.01949-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiner RM, Taylor LE, Henrissat B, Hauser L, Land M, Coutinho PM, Rancurel C, Saunders EH, Longmire AG, Zhang H, et al. Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40 T. PLoS Genet. 2008;4:e1000087. doi: 10.1371/journal.pgen.1000087. 10.1371/journal.pgen.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson IJ, Dharmarajan L, Rodriguez J, Hooper S, Porat I, Ulrich LE, Elkins JG, Mavromatis K, Sun H, Land M, et al. The complete genome sequence of Staphylothermus marinus reveals differences in sulfur metabolism among heterotrophic Crenarchaeota. BMC Genomics. 2009;10:145. doi: 10.1186/1471-2164-10-145. 10.1186/1471-2164-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson I, Ulrich LE, Lupa B, Susanti D, Porat I, Hooper SD, Lykidis A, Sieprawska-Lupa M, Dharmarajan L, Goltsman E, et al. Genomic characterization of methanomicrobiales reveals three classes of methanogens. PLoS ONE. 2009;4:e5797. doi: 10.1371/journal.pone.0005797. 10.1371/journal.pone.0005797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin M, Zhang C, Gibson K, Rikihisa Y. Analysis of complete genome sequence of Neorickettsia risticii: causative agent of Potomac horse fever. Nucleic Acids Res. 2009;37:6076–6091. doi: 10.1093/nar/gkp642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmann JD, Chamberlin MJ. Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 19.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eddy S. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 21.Galperin MY, Natale DA, Aravind L, Koonin EV. A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol. 1999;1:303–305. [PMC free article] [PubMed] [Google Scholar]
- 22.Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Meth. Enzymol. 2007;422:1–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ermolaeva MD. Operon finding in bacteria. In: Subramaniam S, editor. Encyclopedia of Genetics, Genomics, Proteomics, and Bioinformatics. New York: John Wiley & Sons; 2005. pp. 2886–2891. [Google Scholar]
- 24.Dam P, Olman V, Harris K, Su Z, Xu Y. Operon prediction using both genome-specific and general genomic information. Nucleic Acids Res. 2006;35:288–298. doi: 10.1093/nar/gkl1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pertea M, Ayanbule K, Smedinghoff M, Salzberg SL. OperonDB: a comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res. 2008;37:D479–D482. doi: 10.1093/nar/gkn784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 27.Dutta S, Burkhardt K, Young J, Swaminathan GJ, Matsuura T, Henrick K, Nakamura H, Berman HB. Data deposition and annotation at the worldwide protein data bank. Mol. Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- 28.The UniProt Consortium. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koretke KK, Lupas AN, Warren PV, Rosenberg M, Brown JR. Evolution of two-component signal transduction. Mol. Biol. Evol. 2000;17:1956–1970. doi: 10.1093/oxfordjournals.molbev.a026297. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 32.van Dongen S. A cluster algorithm for graphs. Inf. Syst. 2000;1:1–40. [Google Scholar]