Abstract
Most human diseases are complex multi-factorial diseases resulting from the combination of various genetic and environmental factors. In the KEGG database resource (http://www.genome.jp/kegg/), diseases are viewed as perturbed states of the molecular system, and drugs as perturbants to the molecular system. Disease information is computerized in two forms: pathway maps and gene/molecule lists. The KEGG PATHWAY database contains pathway maps for the molecular systems in both normal and perturbed states. In the KEGG DISEASE database, each disease is represented by a list of known disease genes, any known environmental factors at the molecular level, diagnostic markers and therapeutic drugs, which may reflect the underlying molecular system. The KEGG DRUG database contains chemical structures and/or chemical components of all drugs in Japan, including crude drugs and TCM (Traditional Chinese Medicine) formulas, and drugs in the USA and Europe. This database also captures knowledge about two types of molecular networks: the interaction network with target molecules, metabolizing enzymes, other drugs, etc. and the chemical structure transformation network in the history of drug development. The new disease/drug information resource named KEGG MEDICUS can be used as a reference knowledge base for computational analysis of molecular networks, especially, by integrating large-scale experimental datasets.
INTRODUCTION
Twenty years ago, the Human Genome Project was initiated aiming to uncover the genetic factors of human diseases and to develop new strategies for diagnosis, treatment and prevention. The successful sequencing of the human genome and the following coordinated efforts, such as the HapMap project (1), genome-wide association studies (2) and the cancer genome projects (3), have resulted in the discovery of many disease-associated genes. However, our understanding of molecular mechanisms is still largely incomplete for the majority of diseases, which are multi-factorial diseases resulting from the combination of various genetic and environmental factors. There must be inherent relationships among these factors for the etiology and pathogenesis, and they may be characterized by considering the molecular networks involving these factors. The analysis of network–disease associations, in addition to gene–disease associations, would better clarify the molecular mechanisms of diseases and help develop new drugs and treatments.
The KEGG database project was initiated in 1995, originally as part of the Japanese Human Genome Program (4). Since then we have been organizing our knowledge of cellular functions and organism behaviors in computable forms, especially in the forms of molecular networks (KEGG pathway maps) and hierarchical lists (BRITE functional hierarchies). This computerized knowledge has been widely used as a reference for biological interpretation of large-scale datasets generated by sequencing and other high-throughput experimental technologies. Our efforts are now focused on human diseases and drugs. We consider diseases as perturbed states of the molecular system that operates the cell and the organism, and drugs as perturbants to the molecular system. There are a number of disease databases available (5), but they are mostly descriptive databases for humans to read and understand. In KEGG, disease information is being organized in more computable forms: pathway maps and gene/molecule lists. Here, we summarize the current status of the KEGG project and the new developments of the KEGG DISEASE and KEGG DRUG databases that comprise KEGG MEDICUS.
OVERVIEW OF KEGG
Molecular building blocks
KEGG (Kyoto Encyclopedia of Genes and Genomes) is an integrated database resource consisting of 16 main databases, broadly categorized into systems information, genomic information and chemical information as shown in Table 1. Genomic and chemical information represents the molecular building blocks of life in the genomic and chemical spaces, respectively, and systems information represents functional aspects of the biological systems, such as the cell and the organism, that are built from the building blocks. The actual data contents of the genomic information category are: gene catalogs (KEGG GENES) in the completely sequenced genomes (KEGG GENOME), computationally derived sequence similarity relationships (KEGG SSDB), manually defined ortholog groups (KEGG ORTHOLOGY) and supplementary gene catalog data (KEGG DGENES and EGENES). The data contents of the chemical information category are: small molecules (KEGG COMPOUND), glycans (KEGG GLYCAN), reactions among them (KEGG REACTION), chemical structure transformation patterns derived from them (KEGG RPAIR) and supplementary information on enzyme nomenclature (KEGG ENZYME). The five databases in the chemical information category are collectively called KEGG LIGAND. Note that KEGG DRUG has been moved from the chemical information category (6) to the systems information category to integrate with KEGG DISEASE.
Table 1.
KEGG databases
| Category | Database | Content |
|---|---|---|
| Systems Information | KEGG PATHWAY | Pathway maps |
| KEGG BRITE | Functional hierarchies | |
| KEGG MODULE | Pathway modules | |
| KEGG DISEASE | Human diseases | |
| KEGG DRUG | Drugs | |
| Genomic Information | KEGG ORTHOLOGY | KEGG orthology (KO) groups |
| KEGG GENOME | KEGG organisms | |
| KEGG GENES | Genes in high-quality genomes | |
| KEGG SSDB | Sequence similarities and best hit relations | |
| KEGG DGENES | Genes in draft genomes | |
| KEGG EGENES | Genes as EST contigs | |
| Chemical Information | KEGG COMPOUND | Metabolites and other small molecules |
| KEGG GLYCAN | Glycans | |
| KEGG REACTION | Biochemical reactions | |
| KEGG RPAIR | Reactant pair chemical transformations | |
| KEGG ENZYME | Enzyme nomenclature |
Molecular systems
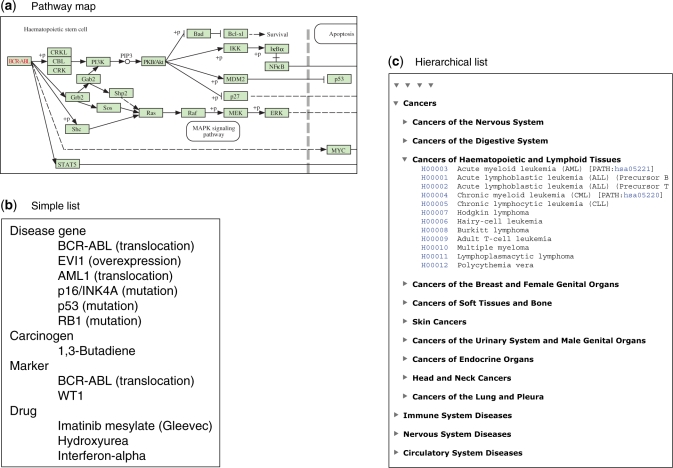
The systems information category is the most unique feature in the KEGG resource. Data and knowledge about the molecular systems that govern cellular processes and organism behaviors are manually collected and summarized from literature and presented in computable forms, called ‘pathway map’ (graph), ‘simple list’ (membership) and ‘hierarchical list’ (ontology), as illustrated in Figure 1. The pathway map in KEGG PATHWAY represents our knowledge about various types of molecular networks: reaction/interaction networks for metabolism, genetic information processing, environmental information processing and other cellular processes; perturbed reaction/interaction networks for human diseases; and relation networks (chemical structure transformation networks) for drug development. The molecular network shown in the pathway map is a graph consisting of nodes (orthologs, genes, proteins, small molecules, etc.) and edges (reactions, interactions and relations). In contrast, the simple list is a set of nodes without the wiring information, simply representing the membership to a molecular system. This representation is used when the detail of the molecular system is not known (KEGG DISEASE) or when a tighter functional unit is indicated within the molecular network (KEGG MODULE). The hierarchical list in KEGG BRITE represents hierarchically organized membership information, which may be called ontology. It is used to represent our knowledge on functional hierarchies inherent in various types of biological systems; not only molecular systems, but also cellular and organismal systems. The content of KEGG BRITE is currently classified into: genes and proteins; compounds and reactions; drugs and diseases; and cells and organisms.
Figure 1.
Knowledge representation of systemic functions in the forms of (a) pathway map; (b) simple list; and (c) hierarchical list. Examples shown here are: (a) KEGG PATHWAY entry (hsa05220) for chronic myeloid leukemia, KEGG DISEASE entry (H00004) for chronic myeloid leukemia and KEGG BRITE entry (br08402) for a classification of human diseases.
NEW DEVELOPMENTS IN KEGG
KEGG GENES and ortholog annotation
One of the main objectives of the KEGG project has been to uncover higher level systemic functions of the cell and the organism from genomic and molecular-level information. The basis for genome annotation in KEGG, which is continuously performed for all sequenced genomes, is the KO system consisting of manually defined ortholog groups that correspond to individual nodes in the KEGG pathway maps and the BRITE functional hierarchies. Once genes are assigned KO identifiers or K numbers by the ortholog annotation procedure described below, the collective body of K numbers can be mapped to KEGG pathway maps and BRITE functional hierarchies, highlighting any subsystems present and enabling higher level functional interpretation of the genome.
During the past two years, the ortholog annotation procedure has been significantly improved by the newly developed KOALA (KEGG Orthology and Links Annotation) tool. There are two types of annotation in KEGG. One is a genome-based annotation, assigning K numbers to genes in a given genome. The other is a KO-based annotation, assigning a given K number (such as in a pathway map) to genes in all organisms. In order to cope with an increasing number of complete genomes, the first annotation is now partially automated (except for selected reference organisms) with continuous efforts to manually improve the second cross-species annotation. The current KEGG annotation procedure is as follows.
Gene information for completely sequenced genomes is computationally generated from RefSeq (7) and other public resources, and stored in the KEGG GENES database.
Sequence similarity scores and best-hit relations are computationally generated from KEGG GENES by pair-wise genome comparisons using SSEARCH, and stored in the KEGG SSDB database.
Automatic genome-based annotation is performed for a limited set (currently, about one-third) of K numbers, which are considered safe for such purpose based on the result of SSDB computation and the criteria of the KOALA tool.
Manual annotation is performed across species for other K numbers using the KOALA and GFIT (8) tools. This step may involve addition/revision of ortholog groups, which is essential to increase the number of safe K numbers.
A glimpse of this procedure can be seen through the read-only versions of KOALA and GFIT tools available on the KO and GENES entry pages, respectively. The quality of KEGG ortholog annotation can be examined by two additional tools. The ortholog table tool displays the status of KO assignment for a given set of K numbers, which is useful to check the completeness of a pathway or a complex. The gene cluster tool displays the status of KO assignment along the chromosomal position of a given genome, which is useful to check the consistency of annotation for operon-like structures in bacterial genomes.
As of 3 September 2009, the KEGG GENES database contains 4.8 million genes in 1049 genomes. In comparison, the UniProt database (9) contains 9.4 million proteins from one-half million species. KEGG already covers half of the known protein universe and >90% of protein sequence families (Kanehisa,M., unpublished data). As the number of complete genomes increases, the coverage of the protein universe will also increase, but there will be remaining fractions of protein families, such as for plant proteins and viral proteins. These protein families are useful to analyze, for example, EST data and metagenomics data, and they will be incorporated in the KO system.
KEGG PATHWAY and BRITE: reference knowledge bases
The KEGG reference pathway maps and BRITE reference hierarchies are created in a general way to be applicable to all organisms; namely, in terms of the orthologs defined by K numbers. The organism-specific pathways and hierarchies can then be generated by converting K numbers to gene identifiers in a given organism. In the past year, the KEGG PATHWAY database has been completely renovated. All the pathway maps have been redrawn using a newly developed tool called KegSketch, which generates KGML+ (meaning KGML + SVG) files. Internally the database update procedure is now based on the text manipulation of these files rather than the color manipulation of image files. For outside services, the coloring procedure continues to be done on image files, but the image file format has been changed from GIF to PNG to accommodate more colors. As a result, there is now no distinction between the global map (6) and the regular pathway maps; they can be manipulated in the same way both in the new KEGG Atlas tool and the traditional image map viewer. Another new feature in the outside service is the XML version of KEGG pathway maps, which is made available in both the original KGML format and the converted BioPAX level 2 format (10).
KEGG LIGAND for chemical bioinformatics
The KEGG LIGAND database contains information about chemical structures and chemical reactions of endogenous molecules, small molecules to larger biopolymers. Certain KEGG pathway maps contain reference chemical structures that can be used to link genomes to the chemical diversity of endogenous molecules. For example, the KEGG pathway map for N-glycan biosynthesis (map00510) contains both the biosynthetic pathway and the synthesized glycan structure. By mapping the genomic content of glycosyltransferases, such as for human (hsa00510), the organism-specific pathway and the organism-specific glycan structure can be seen. This type of structural mapping has been done more extensively in eukaryotic genomes to characterize the chemical structural diversity of glycans (11) and lipids (12). A potentially more interesting, but more difficult, problem is to link plant genomes to plant secondary metabolites. Plants are known to produce diverse chemical compounds including those with medicinal and nutritional values, but the chemical architecture is more complex than simple biopolymers of glycans and lipids. We have introduced KEGG PLANT, a new interface to the KEGG resource for plant research, especially for understanding relationships between genomic and chemical information of plant natural products.
We have also been trying to expand our knowledge on biochemical reactions from experimentally characterized reactions in Enzyme Nomenclature (KEGG ENZYME) to pathway-based definition of reactions (KEGG REACTION) to chemical structural motifs, called RDM patterns that characterize reactions (KEGG RPAIR). The RDM patterns have been used to predict microbial biodegradation pathways from chemical structures of environmental compounds (13). As an extension of this line of research, the E-zyme tool for reaction prediction from a pair (or pairs) of chemical structures has been upgraded by introducing a new algorithm (14).
KEGG MEDICUS for analysis of network–disease associations
In KEGG, disease and drug information is being organized in more computable forms, especially for the analysis of molecular networks. As shown in Table 2, the disease/drug resource, called KEGG MEDICUS, consists of each of the KEGG DISEASE and KEGG DRUG databases, a specific category of the KEGG PATHWAY database and a specific category of the KEGG BRITE database. Disease information is computerized in two forms: pathway maps and gene/molecule lists. The Human Diseases category of the KEGG PATHWAY database contains about 40 pathway maps for cancers, immune disorders, neurodegenerative diseases, circulatory diseases, metabolic disorders and infectious diseases. When the detail of the molecular network is not known but disease genes are identified, we use the gene/molecule list representation and create a KEGG DISEASE entry. The entry contains a list of known disease genes and other relevant molecules including environmental factors, diagnostic markers and therapeutic drugs. The list simply defines the membership to the underlying molecular system, but is still useful for computational analysis.
Table 2.
KEGG MEDICUS for disease and drug information
| KEGG DISEASE | KEGG DRUG | |
|---|---|---|
| URL | http://www.genome.jp/kegg/disease/ | http://www.genome.jp/kegg/drug/ |
| Database | Gene/molecule lists consisting of disease genes, environmental factors, diagnostic markers, and therapeutic drugs | Chemical structures and/or components of approved drugs in Japan, United States and Europe with additional information including: targets, metabolizing enzymes and drug interactions |
| Pathway map | KEGG pathway maps for human diseases including: cancers, immune disorders, neurodegenerative diseases, circulatory diseases, metabolic disorders and infectious diseases | KEGG pathway maps for drug metabolism; KEGG pathway maps for biosynthesis of antibiotics and natural products; and KEGG DRUG structure maps for drug development |
| BRITE hierarchy | Disease classifications including: Pathogens and infectious diseases; Human diseases; and ICD-10 disease classification | Drug classifications including: Therapeutic category of drugs (Japan); ATC classification (WHO); TCM (Traditional Chinese Medicine) drugs in Japan; and Crude drugs in Japan |
| Release | 2008 | 2005 |
The KEGG DRUG database is a chemical structure-based information resource for all prescription and OTC drugs in Japan including crude drugs and TCM formulas, as well as most prescription drugs in the USA and many prescription drugs from Europe. In addition to chemical structures (or chemical compositions for multi-component drugs) and therapeutic efficacy of about 9000 drugs (as of September 2009), different drug classification systems are maintained as part of the KEGG BRITE functional hierarchies. Some are based on the established classification systems to which KEGG DRUG entries are assigned, including the ATC (Anatomical Therapeutic Chemical) classification by WHO, therapeutic category of prescription drugs in Japan and classification of OTC drugs in Japan. There are additional classification systems developed by KEGG, including those for crude drugs and TCM formulas.
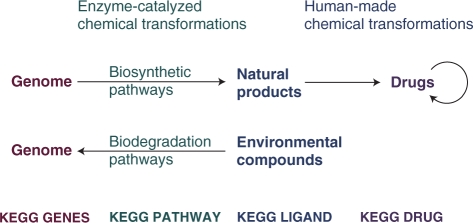
Furthermore, KEGG DRUG contains information about two types of molecular networks. The first network is a molecular interaction network representing interactions and/or relations with target molecules (often in the context of pathway maps), drug metabolizing enzymes, drug transporters and other drugs (especially those causing adverse effects). The second network is a network of chemical structure changes in small molecules, which includes series of chemical modifications introduced by medicinal chemists in the history of drug development (in the KEGG drug structure maps), secondary metabolic pathways for biosynthesis of druggable natural products and drug metabolism (both in the KEGG pathway maps). We have analyzed the chemical architecture of marketed drugs and the patterns of chemical structure transformations in the history of drug development (15), in a similar spirit to the RDM patterns of chemical structure transformations in enzyme-catalyzed reactions. As illustrated in Figure 2, the second network may be used to analyze the chemical architecture of natural products and the chemical architecture of marketed drugs towards drug discovery from the genomes of plants and microorganisms. Furthermore, the second network may have relevance in understanding drug metabolism and biodegradation of environmental substances by considering not only the human genome but also the metagenome of the human body.
Figure 2.
KEGG accumulates knowledge about the networks of chemical structure transformations for linking genomes to chemical structures.
ACCESSING KEGG
Web sites
KEGG is made available at both the GenomeNet web site (http://www.genome.jp/kegg/) and the KEGG website (http://www.kegg.jp/). The KEGG Atlas tool is available only at the KEGG web site.
KEGG identifiers
Each database entry in KEGG is identified by the form of ‘db:entry’ where ‘db’ is the database name and ‘entry’ is the entry identifier. With the new version of the DBGET database retrieval system (16) the db prefix is no longer necessary, because the entry identifiers are unique across all the KEGG databases. For example, C00022 is sufficient rather than cpd:C00022. See more details at the KEGG identifiers page (http://www.genome.jp/kegg/kegg3.html). This simple form can be used to retrieve individual database entries, pathway maps and BRITE hierarchies by the REST-based URLs (http://www.genome.jp/kegg/docs/weblink.html) at the KEGG web site.
FUNDING
Institute for Bioinformatics Research and Development of the Japan Science and Technology Agency (to KEGG project); Grant-in-aid for scientific research on the priority area ‘Comprehensive Genomics’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to KEGG project). Funding for open access charge: grant-in-aid for scientific research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The computational resource was provided by the Bioinformatics Center, Institute for Chemical Research, Kyoto University.
REFERENCES
- 1.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 2.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 3.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanehisa M. A database for post-genome analysis. Trends Genet. 1997;13:375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- 5.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The UniProt Consortium. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Küntzer J, Backes C, Blum T, Gerasch A, Kaufmann M, Kohlbacher O, Lenhof HP. BNDB – the Biochemical Network Database. BMC Bioinformatics. 2007;8:367. doi: 10.1186/1471-2105-8-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto K, Tokimatsu T, Kawano S, Yoshizawa AC, Okuda S, Goto S, Kanehisa M. Comprehensive analysis of glycosyltransferases in eukaryotic genomes for structural and functional characterization of glycans. Carbohydr. Res. 2009;344:881–887. doi: 10.1016/j.carres.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto K, Yoshizawa AC, Okuda S, Kuma K, Goto S, Kanehisa M. The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J. Lipid Res. 2008;49:183–191. doi: 10.1194/jlr.M700377-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Oh M, Yamada T, Hattori M, Goto S, Kanehisa M. Systematic analysis of enzyme-catalyzed reaction patterns and prediction of microbial biodegradation pathways. J. Chem. Inf. Model. 2007;47:1702–1712. doi: 10.1021/ci700006f. [DOI] [PubMed] [Google Scholar]
- 14.Yamanishi Y, Hattori M, Kotera M, Goto S, Kanehisa M. E-zyme: predicting potential EC numbers from the chemical transformation pattern of substrate-product pairs. Bioinformatics. 2009;25:i79–i86. doi: 10.1093/bioinformatics/btp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shigemizu D, Araki M, Okuda S, Goto S, Kanehisa M. Extraction and analysis of chemical modification patterns in drug development. J. Chem. Inf. Model. 2009;49:1122–1129. doi: 10.1021/ci8003804. [DOI] [PubMed] [Google Scholar]
- 16.Kanehisa M. Linking databases and organisms - GenomeNet resources in Japan. Trends Biochem. Sci. 1997;22:442–444. doi: 10.1016/s0968-0004(97)01130-4. [DOI] [PubMed] [Google Scholar]