Abstract
CORUM is a database that provides a manually curated repository of experimentally characterized protein complexes from mammalian organisms, mainly human (64%), mouse (16%) and rat (12%). Protein complexes are key molecular entities that integrate multiple gene products to perform cellular functions. The new CORUM 2.0 release encompasses 2837 protein complexes offering the largest and most comprehensive publicly available dataset of mammalian protein complexes. The CORUM dataset is built from 3198 different genes, representing ∼16% of the protein coding genes in humans. Each protein complex is described by a protein complex name, subunit composition, function as well as the literature reference that characterizes the respective protein complex. Recent developments include mapping of functional annotation to Gene Ontology terms as well as cross-references to Entrez Gene identifiers. In addition, a ‘Phylogenetic Conservation’ analysis tool was implemented that analyses the potential occurrence of orthologous protein complex subunits in mammals and other selected groups of organisms. This allows one to predict the occurrence of protein complexes in different phylogenetic groups. CORUM is freely accessible at (http://mips.helmholtz-muenchen.de/genre/proj/corum/index.html).
INTRODUCTION
Major cellular processes like cell cycle, protein folding and protein degradation depend on the activity of protein complexes (1). To date there are no reliable estimates about the total number of protein complexes in cells (complexome), but data from single cell organisms provide evidence, that more than half of the gene products are involved in the formation of protein complexes (2). In the advent of protein network analyses, topological properties of protein complexes resulted in paraphrases such as ‘party hubs’ (3) or ‘multi-interface hubs’ (4). Bioinformatics analysis of protein–protein interaction (PPI) datasets revealed that protein complex subunits are stronger evolutionary conserved and show a higher essentiality than proteins from other interactions (4).
As the most comprehensive PPI and protein complex data are available for Saccharomyces cerevisiae, most of these discoveries were obtained using data from yeast. In addition to a manually curated dataset of protein complexes (5), tag-based high-throughput approaches were performed in order to define the yeast complexome (6,7). The importance of manually curated gold-standards was demonstrated by analyses of results from high-throughput experiments. In an assessment of different high-throughput technologies for the analysis of PPIs it was shown, that each method, depending on its physiochemical constraints, captures interactions for different subsets of proteins (8). Thus, none of the existing methods is able to detect all interactions and it was also shown that even the combined dataset of five different methods missed ∼40% of experimentally validated, manually curated interactions (9).
For mammals no comprehensive high-throughput dataset of protein complexes is publicly available. Bioinformatics analyses of the mammalian complexome can be performed either by using artificially constructed protein complexes (10) or data from manually curated datasets (11,12). In 2008, the CORUM database was introduced as the most comprehensive catalogue of mammalian protein complexes. All data are manually curated including information of protein complex subunits and methods of purification as well as additional information such as functional annotation using the Functional Catalogue (FunCat) annotation scheme (13), stoichiometry of the subunits and information about association with diseases (14). Analyses of the CORUM dataset have shown (i) that mammalian protein complexes are most frequently composed of 3 or 4 different subunits and (ii) that proteins tend to be reused in up to 53 protein complexes (15).
The CORUM dataset has been used for a number of bioinformatics analyses like tissue-specific expression of proteins (16), functional interpretation of high-throughput data (17–19) or to predict interactions of protein regions (20). In addition, the dataset contributes to web-based applications like the DICS database of functional modules (21) or the COFECO tool for composite function annotation (22).
The CORUM Release 2.0 presents a significantly extended dataset that now consists of 2837 mammalian protein complexes. In addition to existing cross-references the dataset was mapped to Entrez Gene identifiers and functional annotation of Gene Ontology (GO) terms. In order to enable more specific search results in comments, the content is now distributed into the three sections ‘Disease Comment’, ‘Functional Comment’ and ‘Subunit Comment’. Finally, an analysis tool was implemented that allows one to predict the occurrence of orthologous protein complex subunits in other mammals and other groups of organisms. The ‘Phylogenetic Conservation’ tool provides a probability whether or not a protein complex is likely to occur in the analysed model organisms. CORUM is freely accessible at http://mips.helmholtz-muenchen.de/genre/proj/corum/index.html.
NEW DEVELOPMENTS
Dataset and cross-references
In 2008 the CORUM dataset consisted of 1750 mammalian protein complexes, mainly characterized in human (60%), mouse (14%) and rat (14%) (14). While the relative abundance of the related organisms remained stable in the meantime, the number of protein complexes has grown to 2837 in September 2009. Thus, CORUM is the largest set of mammalian protein complexes publicly available.
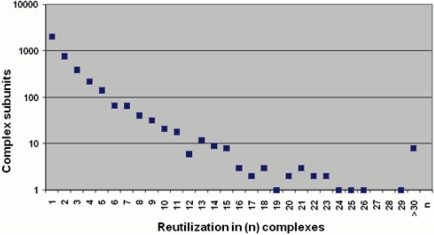
However, compared to data from single-cell organisms only a minor fraction of the mammalian complexome has been discovered so far. Data from yeast have shown that at least 45% of the gene complement function as subunits in protein complexes (14). Considering that there is no comprehensive mammalian high-throughput dataset available to date, the fraction of genes that are involved in protein complex formation is comparably low. These estimates are based on the number of different complex subunit genes divided by a given number of 20 488 genes in human (14). Compared to the first CORUM release, this fraction increased moderately from 12% (2400 genes) to 16% (3198 genes). The slow increase of novel protein complex subunits presumably results from the reuse of subunits (Figure 1) in different protein complexes or protein complex variants (15). Data from the CORUM ‘Core Set’ (see below) show that proteins like ‘integrin beta-1’, ‘histone deacetylase 1’ and ‘histone deacetylase 2’ appear in 54, 51 and 38 different human protein complexes. Multiple reutilization of protein complex subunits is particularly found in large protein complex families like SNARE complexes and ubiquitin E3 ligases. The ubiquitin E3 ligase subunit ring-box 1 (Rbx1), for example, was identified in 35 complexes.
Figure 1.
Reutilization of protein complex subunits. The plot shows that most proteins (2038) are found in only one protein complex and only eight proteins are subunits of at least 30 protein complexes. Data for the analysis are based on the CORUM ‘Core Set’.
In addition to the complete dataset, CORUM now offers a reduced ‘Core Dataset’ for download and searches that avoids redundancies of data. Thoroughly investigated protein complexes like ‘SNARE complex (Vamp2, Snap25, Stx1a, Cplx1)’, ‘succinyl-CoA synthetase, ADP-forming’ and ‘cytochrome bc1-complex (EC 1.10.2.2), mitochondrial’ are characterized in more than one mammalian organism. Due to the close phylogenetic relationship between mammals it can be assumed that the majority of protein complexes are conserved in mammals. However, as the aim of CORUM is to provide a comprehensive dataset, also evolutionary conserved protein complexes from different organisms (interologous protein complexes) are annotated in CORUM. To some extent this introduces redundancies, but on the other hand proves that the same protein complex in fact exists in different organisms.
Results from several laboratories that investigated the same protein complex but characterized the molecule with a different composition are another source of dataset expansion. These may stem from different experimental conditions that result in different complex compositions depending on the stringency of the experimental procedures or from different biomaterial that was used for the characterization. Bioinformatics applications like machine learning require non-redundant datasets. For these users we offer the ‘Core Set’ of 2084 distinct protein complexes. For the set only one representative of each interologous group of protein complexes or from protein complex variants was selected. We chose protein complexes which were thoroughly characterized and preferably from Homo sapiens.
Annotation of protein complex subunits in CORUM is performed with UniProt identifiers. Since some users prefer identifiers from Entrez Gene, we mapped the UniProt identifiers to the corresponding Entrez Gene identifiers. This was realized in a semi-automatic procedure using the CRONOS tool (23). CRONOS allows the mapping of identifiers, gene names and protein names from various resources like UniProt, RefSeq and Ensembl. In total, 4310 out of 4336 distinct subunits (98%) could be mapped to corresponding Entrez Gene identifiers. For 26 gene products like MRPS15 from Bos taurus or SPCS1 from Canis familiaris no respective identifier was available in Entrez.
CORUM is the only resource of protein complexes that includes functional annotation of the molecules. We use the FunCat annotation scheme for protein and protein complex function characterization (13). The FunCat has been used for genome annotation and was also frequently used for the analysis of protein networks and high-throughput experiments (13). The hierarchical structure of the FunCat allows browsing for protein complexes with particular cellular functions or localizations. In recent years, GO has become a widely used tool for the annotation of eukaryotic genomes (24). In contrast to the FunCat annotation scheme, the GO is constructed as a set of acyclic graphs, allowing more than one parent class per child (24). In order to enable bioinformatics analyses of protein complexes based on GO terms, the new CORUM release provides a mapping from FunCat to GO. The mapping was performed using the table that is available for download at http://www.geneontology.org/external2go/mips2go. As a result 840 FunCat categories could be mapped to 896 GO terms. Manual inspection of 100 randomly chosen protein complexes revealed that FunCat categories and GO terms are in agreement.
Some valuable information concerning protein complexes cannot be covered by systematic annotation schemes but is represented as free text comment in CORUM. This information includes protein complex composition (e.g. additional subunits of unknown identity), association of protein complexes with diseases or particular functional properties. In the first CORUM release this additional information was collected in a single comment field. In CORUM release 2.0 this content is now distributed among the three comment fields ‘Functional Comment’, ‘Disease Comment’ and ‘Subunit Comment’. This separation allows to search in a particular type of information or using a wild card ‘_’ for instance to retrieve all 223 protein complexes with information about disease association.
Phylogenetic analysis of protein complexes
Protein complex subunits from protein complexes like ribosomes and chaperonins are highly conserved in evolution. Beside ribosomal RNAs, subunits from complexes such as RNA polymerases (25) and F1-ATPases (26) were used for phylogenetic analyses in the early days of sequence-based phylogenetic analyses. Based on data from 191 sequenced genomes, 2 years ago a novel endeavor was started to investigate highly conserved proteins for phylogenetic analysis (27). Analysis revealed 31 highly conserved proteins that allow a new reconstruction of the tree of life and 28 of these proteins are known to be protein complex subunits (23 ribosomal proteins). To enable scientists to obtain some insight into the phylogenetic conservation of subunits, the ‘Phylogenetic Conservation’ tool has been developed for comparative proteome analysis. The ‘Phylogenetic Conservation’ tool is based on sequence similarity data that are obtained from the SIMAP database (28). The Similarity Matrix of Proteins (SIMAP) database provides a comprehensive and up-to-date dataset of the pre-calculated sequence similarity matrix and sequence-based features like InterPro domains for all proteins contained in the major public sequence databases.
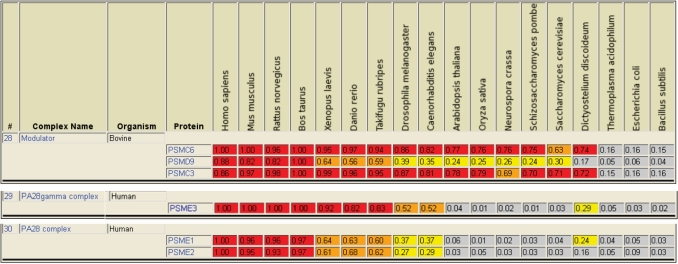
The ‘Phylogenetic Conservation’ tool in CORUM presents the similarity of the protein complex subunits to proteins from other organisms as tables (Figure 2). As default comparison to 18 organisms are shown, four mammals (Homo sapiens, Mus musculus, Rattus norvegicus and Bos taurus), three other vertebrates (Xenopus laevis, Danio rerio and Takifugu rubripes), two invertebrates (Caenorhabditis elegans and Drosophila melanogaster), two plants (Arabidopsis thaliana and Oryza sativa), three fungi (Neurospora crassa, Schizosaccharomyces pombae and S. cerevisiae), one slime mold (Dictyostelium discoideum) and three prokaryotes (Thermoplasma acidophilum, Escherichia coli and Bacillus subtilis). In addition to the numerical values, the degree of protein sequence similarity is colour coded.
Figure 2.
Phylogenetic conservation of proteasome regulatory protein complexes. Results of the phylogenetic conservation tool from CORUM for the three proteasome regulators ‘Modulator’, ‘PA28 gamma complex’ and ‘PA28 complex’ are shown. Similarity of protein complex subunits to proteins from other organisms are represented color coded as well as opt. score/self score ratios. The data are obtained from the SIMAP database.
The conservation of protein complexes appears to be conserved among all phylogenetic related organisms and separates organisms of distant phylogenetic relation, depending on the respective complex. This can be illustrated with the proteasome and three proteasome activatory complexes. Two subunits of the ‘Modulator (PA700-dependent proteasome activator)’ are highly conserved (red colour) within all eukaryotes, whereas the ‘PA28 gamma complex’ is only highly conserved within vertebrates (Figure 2). Finally, high conservation of the ‘11 S REG complex’ is restricted to the four mammalian proteomes. The 20 S proteasome complex is a high-molecular-weight protease that is essential for protein degradation in mammals. Results of the ‘Phylogenetic Conservation’ tool reveal weak similarity for proteins in the archaeon T. acidophilum (Supplementary Figure S1). In fact, an archetype of proteasomes, consisting of only two different subunits is frequently found in archaea (29). On the other hand, sophisticated proteasome architectures like the 26 S proteasome or the availability of several proteasome activatory complexes are not found in Thermoplasma or other prokaryotes. In agreement with this observation, the three above mentioned proteasome activators show no similarity to proteins from Thermoplasma (Figure 2). Results of the ‘Phylogenetic Conservation’ tool can be retrieved for single protein complexes or for multiple complexes that were found by one of the search options in CORUM.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
ERA-NET PathoGenoMics ‘Pathomics’ grant (BMBF) (to B.W.). Funding to open access charge: Helmholtz Center Munich (Helmholtz Zentrum München).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Thomas Rattei for providing data from SIMAP.
REFERENCES
- 1.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Guldener U, Munsterkotter M, Kastenmuller G, Strack N, Van Helden J, Lemer C, Richelles J, Wodak SJ, Garcia-Martinez J, Perez-Ortin JE, et al. CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 2005;33:D364–D368. doi: 10.1093/nar/gki053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 4.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 5.Guldener U, Munsterkotter M, Oesterheld M, Pagel P, Ruepp A, Mewes HW, Stumpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 7.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 8.Jensen LJ, Bork P. Biochemistry. Not comparable, but complementary. Science. 2008;322:56–57. doi: 10.1126/science.1164801. [DOI] [PubMed] [Google Scholar]
- 9.Braun P, Tasan M, Dreze M, Barrios-Rodiles M, Lemmens I, Yu H, Sahalie JM, Murray RR, Roncari L, de Smet AS, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tumer Z, Pociot F, Tommerup N, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 11.Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, et al. Human protein reference database—2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, Waegele B, Schmidt T, Doudieu ON, Stumpflen V, et al. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2008;36:D646–D650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong P, Althammer S, Hildebrand A, Kirschner A, Pagel P, Geissler B, Smialowski P, Blochl F, Oesterheld M, Schmidt T, et al. An evolutionary and structural characterization of mammalian protein complex organization. BMC Genomics. 2008;9:629. doi: 10.1186/1471-2164-9-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bossi A, Lehner B. Tissue specificity and the human protein interaction network. Mol. Syst. Biol. 2009;5:260. doi: 10.1038/msb.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedel CC, Dolken L, Ruzsics Z, Koszinowski H, Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37:e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer E, Aglyamova GV, Wang S, Buchanan-Carter J, Abrego D, Colbourne JK, Willis BL, Matz MV. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics. 2009;10:219. doi: 10.1186/1471-2164-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zampieri M, Soranzo N, Altafini C. Discerning static and causal interactions in genome-wide reverse engineering problems. Bioinformatics. 2008;24:1510–1515. doi: 10.1093/bioinformatics/btn220. [DOI] [PubMed] [Google Scholar]
- 20.Schelhorn SE, Lengauer T, Albrecht M. An integrative approach for predicting interactions of protein regions. Bioinformatics. 2008;24:i35–i41. doi: 10.1093/bioinformatics/btn290. [DOI] [PubMed] [Google Scholar]
- 21.Dietmann S, Georgii E, Antonov A, Tsuda K, Mewes HW. The DICS repository: module-assisted analysis of disease-related gene lists. Bioinformatics. 2009;25:830–831. doi: 10.1093/bioinformatics/btp055. [DOI] [PubMed] [Google Scholar]
- 22.Sun CH, Kim MS, Han Y, Yi GS. COFECO: composite function annotation enriched by protein complex data. Nucleic Acids Res. 2009;37:W350–W355. doi: 10.1093/nar/gkp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waegele B, Dunger-Kaltenbach I, Fobo G, Montrone C, Mewes HW, Ruepp A. CRONOS: the cross-reference navigation server. Bioinformatics. 2009;25:141–143. doi: 10.1093/bioinformatics/btn590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puhler G, Leffers H, Gropp F, Palm P, Klenk HP, Lottspeich F, Garrett RA, Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc. Natl Acad. Sci. USA. 1989;86:4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl Acad Sci. USA. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciccarelli FD, Doerks T, von MC, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 28.Rattei T, Arnold R, Tischler P, Lindner D, Stumpflen V, Mewes HW. SIMAP: the similarity matrix of proteins. Nucleic Acids Res. 2006;34:D252–D256. doi: 10.1093/nar/gkj106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupas A, Zuhl F, Tamura T, Wolf S, Nagy I, De MR, Baumeister W. Eubacterial proteasomes. Mol. Biol. Rep. 1997;24:125–131. doi: 10.1023/a:1006803512761. [DOI] [PubMed] [Google Scholar]