Abstract
Chromosome translocations and gene fusions are frequent events in the human genome and have been found to cause diverse types of tumor. ChimerDB is a knowledgebase of fusion genes identified from bioinformatics analysis of transcript sequences in the GenBank and various other public resources such as the Sanger cancer genome project (CGP), OMIM, PubMed and the Mitelman’s database. In this updated version, we significantly modified the algorithm of identifying fusion transcripts. Specifically, the new algorithm is more sensitive and has detected 2699 fusion transcripts with high confidence. Furthermore, it can identify interchromosomal translocations as well as the intrachromosomal deletions or inversions of large DNA segments. Importantly, results from the analysis of next-generation sequencing data in the short read archives are incorporated as well. We updated and integrated all contents (GenBank, Sanger CGP, OMIM, PubMed publications and the Mitelman’s database), and the user-interface has been improved to support diverse types of searches and to enhance the user convenience especially in browsing PubMed articles. We also developed a new alignment viewer that should facilitate examining reliability of fusion transcripts and inferring functional significance. We expect ChimerDB 2.0, available at http://ercsb.ewha.ac.kr/fusiongene, to be a valuable tool in identifying biomarkers and drug targets.
INTRODUCTION
Fusion genes play important roles in tumorigenesis and cancer progression (1). Perhaps, the best-characterized case, BCR-ABL1 fusion is the cause of the chronic myelogenous leukemia and the target of the anticancer drug, Gleevec (imatinib) (2). Identification of fusion genes thus can lead to the discovery of diagnostic biomarkers and therapeutic targets as well as understanding the molecular basis of tumorigenesis.
Initial studies have concentrated on the hematological cancer in large part due to the sample availability (1,3). Over the last few years, however, there has been significant progress in fusion gene identification in solid tumors. Importantly, Chinnaiyan and colleagues (4–7) reported several cases of gene fusion in prostate cancer identified via integrative analysis of microarray data (TMPRSS2 and ETS transcription factors) and transcriptome resequencing. Soda et al. (8) identified the transforming EML4-ALK fusion gene in nonsmall cell lung cancer (NSCLC) using a function-based screening procedure. A proteomic study of phosphotyrosine kinases also revealed the ROS-ALK fusion in NSCLC cell lines (9). These cases clearly indicate that gene fusions play an important role in cancer development in solid tumors.
Recent progress in next-generation sequencing (NGS) techniques provides a tremendous opportunity for fusion gene discovery. Notably, paired-end sequencing, now a frequent if not standard procedure, compensates for the short read length of NGS techniques (10). Sequencing and analyzing whole genome or transcriptome lead to identification of many chromosomal aberrations including translocations, amplifications and deletions. Short read sequencing strategies were successfully applied to find fusion genes in prostate, lung and breast cancer cell lines (5,11–13).
There have been considerable efforts to make a catalog of fusion genes. The Mitelman’s database and the Sanger cancer genome project (CGP) are the notable examples of collecting fusion genes from literature reports (1,3). The COSMIC and CancerGenes database include other types of chromosomal aberrations such as mutations, amplifications and deletions (14,15). Currently, the Mitelman’s database and the Sanger CGP collection include 150 and 270 gene pairs, respectively, involved in gene fusion events.
Bioinformatics analysis of public transcriptome sequences in the GenBank also provides ample cases of fusion transcript candidates. Fusion genes may be classified into two groups, interchromosomal and intrachromosomal. The former results from fusion between two different chromosomes i.e. translocation and the latter originates from single chromosomes due to deletion, inversion or amplification of large DNA segments. Romani et al. (16) analyzed the mRNA sequences and Hahn et al. (17) analyzed the mRNA and EST sequences to identify fusion transcripts between different chromosomes. Similar data-mining approaches were adopted to construct databases of fusion genes such as ChimerDB (18), HybridDB (19) and TICdb (20) although computational details vary considerably.
ChimerDB is designed to be a knowledgebase of fusion genes that encompass the fusion transcripts identified from bioinformatics analysis of transcript sequences in the GenBank and various public resources such as the Sanger CGP (3), OMIM (21), Mitelman’s database (1) and PubMed. The updated version, ChimerDB 2.0, features (i) algorithm modifications for increased sensitivity, (ii) extensive coverage of recent publications and relevant databases, (iii) analysis of NGS data in the NCBI’s short read archives (SRA) and (iv) the enhanced user interface and the novel alignment viewer to support diverse types of search. ChimerDB 2.0 would be the most extensive catalog of fusion genes and transcripts publically available to date.
IMPLEMENTATIONS
Computational method for transcriptome analysis
The basic strategy is virtually identical to the procedure used in ChimerDB 1.0 where the genomic alignments of transcript sequences were analyzed to identify the fusion transcripts. We will describe the major differences and modifications here with more details provided in the Supplementary data and in the web site documentation.
The most important change is relieving the boundary conditions based on our observation that many reported cases did not satisfy the strict condition that the fusion boundary of the transcript should match the exon boundary. Therefore, we introduced the ‘reliability class’ as a measure of confidence level. We consider the alignment with multiple exons or single exon with matching boundaries as features of reliability. Entry to Class A requires that both head and tail transcripts consist of multiple exons or of single exons with matching boundaries, thus being the most reliable cases. Only one or neither of the head and tail genes satisfying this condition would put a given transcript in Class B or C, respectively.
Another important difference is the introduction of various refinement steps. For example, we removed the entries whose genomic alignments have many hits of comparable qualities in different genomic regions even though these genomic regions are not marked as repeat sequences. This step was necessary to avoid possible complications arising from gene duplication, pseudo-genes and retroposon sequences. In addition, the number of exons was estimated by using the Exonerate program rather than the BLAT alignments (22,23).
The computational pipeline for 454 sequences from the SRA is identical with the EST processing since the sequence length is comparable. Solexa reads are generally too short and we used them just as supporting evidence for the existence of fusion transcripts. Solexa transcriptome reads were aligned using the BWA program (24) against the fusion transcripts to determine if multiple reads cover the fusion point. The alignment of resulting candidates was manually examined.
In this updated version, we also include the fusion transcripts within the same chromosome. The head and tails genes, not being adjacent, should be separated by >1 Mb. We exclude the fusion cases between adjacent genes, which we named co-transcription and intergenic splicing, in order to limit our focus to genuine fusion genes originating from chromosomal aberrations.
Data sources
Transcript sequences were downloaded from the GenBank last updated on September, 2008. It included 323 914 mRNA and over 8 million EST sequences for human. We also downloaded NGS transcriptome sequences in the SRA that included ∼1.2 million 454 sequences and ∼762 million Solexa reads. The human genome map used for transcriptome analysis was the NCBI build 36.1 (hg18 in the UCSC genome browser database) (25).
Literature-related information was obtained as follows. PubMed articles related to fusion genes were retrieved by using the Entrez query of ‘chromosomal translocation or fusion gene’ and the MeSH terms on human cancers. Abstracts of 3618 articles were manually examined to obtain information on the fusion gene pairs. OMIM records retrieved by the query of ‘translocation or fusion’ (May 2009) were also manually examined to find fusion gene pairs. As for the Sanger CGP data, the ‘cancer gene census’ list released on December 2008 was downloaded from the web site. Mitelman’s database was obtained from the web site for the recurrent chromosome aberrations in cancer (http://cgap.nci.nih.gov/Chromosomes/RecurrentAberrations) as of April 2009. Entries with specific gene symbols for both head and tail genes were retained as part of our literature-related data.
RESULTS
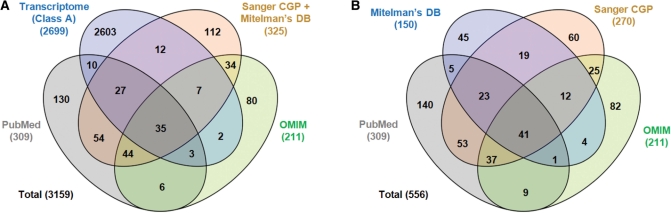
ChimerDB 2.0 includes 9358 genes, 117 47 fusion gene pairs and 9358 fusion transcripts. Figure 1A shows the number of fusion gene pairs according to the information source, counting just the Class A candidates for the transcriptome analysis. As expected, transcriptome analysis is the most ample source of fusion gene pairs and includes 2699 candidates, compared with ∼300 candidates with the original version. Only 96 cases of those have the literature evidence from other resources, implying that the majority of candidates remain to be verified experimentally.
Figure 1.
The number of gene pairs in the ChimerDB 2.0 according to the information source.
Comparison between databases for just the literature-based cases is also revealing (Figure 1B). The overlaps between Sanger CGP, OMIM, Mitelman’s database and our own PubMed collections are much smaller than expected, and 327 cases out of 556 fusion gene pairs are found only in one of the literature databases. This reveals the incomplete coverage of manual efforts and the necessity for integration of various databases. ChimerDB 2.0 includes 537 genes and 556 fusion gene pairs from literature publications, which is a significant increase than other single database.
Detailed statistics of transcriptome analysis is shown in Table 1. We found 1046, 6178 and 2674 fusion transcripts from mRNA, EST and 454 sequences, respectively. In sum, 89% of the total cases are interchromosomal fusions.
Table 1.
Statistics of transcriptome analysis in ChimerDB 2.0
| Class | Interchromosomal |
Intrachromosomal |
||||
|---|---|---|---|---|---|---|
| A | A + B | A + B + C | A | A + B | A + B + C | |
| No. of transcripts | 1900 | 6073 | 8833 | 515 | 887 | 1065 |
| mRNA | 479 | 855 | 900 | 110 | 143 | 146 |
| EST | 1247 | 3972 | 5397 | 396 | 677 | 781 |
| NGS | 174 | 1246 | 2536 | 9 | 67 | 138 |
| No. of genes (9358 in total) | 2855 | 6976 | 8710 | 703 | 1276 | 1543 |
| No. of gene pairs | 2209 | 7362 | 10639 | 490 | 909 | 1108 |
| With multiple transcripts | 278 | 807 | 1137 | 144 | 220 | 246 |
| With Solexa evidence | 14 | 14 | 15 | 65 | 67 | 67 |
A significant proportion of gene pairs (422) in the Class A features multiple fusion transcripts, thus indicating an even higher chance of representing genuine fusion. We also searched the short reads from the Solexa transcriptome sequences in the SRA that span the fusion boundary of our candidates. Notably, 82 fusion transcripts were found to have multiple short-read matches. One of these fusions, CRTC1-MAML2, has been reported to be a frequent feature of mucoepidermoid carcinoma (26). It is noteworthy that intrachromosomal events are overrepresented among our fusion transcript candidates. A close look reveals that a major portion consists of genes belonging to the same family or of pseudogenes. It remains to be seen whether they are from alignment ambiguity or genuine fusion events.
User interface
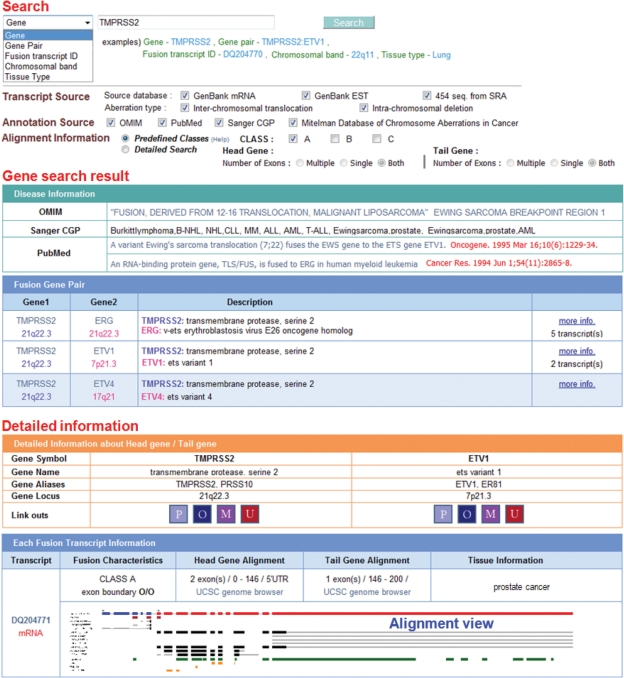
The user interface of ChimerDB is significantly improved in this updated version. Figure 2 shows the important features in the user interface. Most importantly, we support diverse types of search targeting transcripts, genes, gene pairs, cytobands and tissues. As for the fusion transcripts, users may choose the reliability class, number of exons, boundary types for the head and tail genes.
Figure 2.
User interface of ChimerDB 2.0. The search page is designed to support diverse types of search. The ‘search result’ page shows the gene pairs and the disease-related information in OMIM, Sanger CGP and Mitelman’s database with the title and journal name of PubMed articles. Clicking ‘more info’ link shows the detailed information on fusion genes and transcripts as seen in the bottom panel. The ‘alignment view’ shows the hypothetical fusion gene (head gene in blue, tail gene in red) and the candidate fusion transcript (in magenta) along with the UCSC-annotated genes (exons in black, UTRs in grey). The repeat regions and the Pfam domains are indicated in green and orange colors, respectively. Clicking on the alignment picture opens a magnified view. The information contents in this figure are trimmed for brevity.
The result page includes the gene pairs, disease information, PubMed articles and linkouts to diverse resources. PubMed articles are displayed with the title and journal name for user convenience. Except for the few cases without the fusion sequence available, we show the alignment picture that includes the gene structure, domains and repeat sequences. We also provide information on tissue and pathology type from the GenBank records and CGAP (Cancer Genome Anatomy Project) library data. Links to the UCSC genome browser are provided to allow users to examine the detailed gene structure and alignment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Korea Science and Engineering Foundation (KOSEF) funded by the Korea government (MEST) (R01-2008-000-20818-0 and 2007-03983); grant from BioGreen 21 Program of the Korean Rural Development Administration (20070401034010); ‘Systems Biology Infrastructure Establishment Grant’ provided by Gwangju Institute of Science and Technology in 2009 through Ewha Research Center for Systems Biology (ERCSB); grant from the National Core Research Center (NCRC) program (R15-2006-020) of the KOSEF funded by the MEST. Funding for open access charge: Korea Science and Engineering Foundation (R01-2008-000-20818-0).
Conflict of interest statement. None declared.
REFERENCES
- 1.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Treatment for chronic myelogenous leukemia: the long road to imatinib. J. Clin. Invest. 2007;117:2036–2043. doi: 10.1172/JCI31691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat. Rev. Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, et al. Integrative molecular concept modeling of prostate cancer progression. Nat. Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 7.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 8.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 9.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 10.Fullwood MJ, Wei CL, Liu ET, Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521–532. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell PJ, Stephens PJ, Pleasance ED, O'M;eara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan Y, Ooi HS, Choo SW, Chiu KP, Zhao XD, Srinivasan KG, Yao F, Choo CY, Liu J, Ariyaratne P, et al. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using Paired-End diTags (PETs) Genome Res. 2007;17:828–838. doi: 10.1101/gr.6018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Q, Caballero OL, Levy S, Stevenson BJ, Iseli C, de Souza SJ, Galante PA, Busam D, Leversha MA, Chadalavada K, et al. Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line. Proc. Natl Acad. Sci. USA. 2009;106:1886–1891. doi: 10.1073/pnas.0812945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins ME, Claremont M, Major JE, Sander C, Lash AE. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35:D721–D726. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romani A, Guerra E, Trerotola M, Alberti S. Detection and analysis of spliced chimeric mRNAs in sequence databanks. Nucleic Acids Res. 2003;31:e17. doi: 10.1093/nar/gng017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn Y, Bera TK, Gehlhaus K, Kirsch IR, Pastan IH, Lee B. Finding fusion genes resulting from chromosome rearrangement by analyzing the expressed sequence databases. Proc. Natl Acad. Sci. USA. 2004;101:13257–13261. doi: 10.1073/pnas.0405490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim N, Kim P, Nam S, Shin S, Lee S. ChimerDB–a knowledgebase for fusion sequences. Nucleic Acids Res. 2006;34:D21–D24. doi: 10.1093/nar/gkj019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim DS, Huh JW, Kim HS. HYBRIDdb: a database of hybrid genes in the human genome. BMC Genomics. 2007;8:128. doi: 10.1186/1471-2164-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novo FJ, de Mendibil IO, Vizmanos JL. TICdb: a collection of gene-mapped translocation breakpoints in cancer. BMC Genomics. 2007;8:33. doi: 10.1186/1471-2164-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's; Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fehr A, Roser K, Heidorn K, Hallas C, Loning T, Bullerdiek J. A new type of MAML2 fusion in mucoepidermoid carcinoma. Genes Chromosomes Cancer. 2008;47:203–206. doi: 10.1002/gcc.20522. [DOI] [PubMed] [Google Scholar]