Abstract
Linear motifs are short segments of multidomain proteins that provide regulatory functions independently of protein tertiary structure. Much of intracellular signalling passes through protein modifications at linear motifs. Many thousands of linear motif instances, most notably phosphorylation sites, have now been reported. Although clearly very abundant, linear motifs are difficult to predict de novo in protein sequences due to the difficulty of obtaining robust statistical assessments. The ELM resource at http://elm.eu.org/ provides an expanding knowledge base, currently covering 146 known motifs, with annotation that includes >1300 experimentally reported instances. ELM is also an exploratory tool for suggesting new candidates of known linear motifs in proteins of interest. Information about protein domains, protein structure and native disorder, cellular and taxonomic contexts is used to reduce or deprecate false positive matches. Results are graphically displayed in a ‘Bar Code’ format, which also displays known instances from homologous proteins through a novel ‘Instance Mapper’ protocol based on PHI-BLAST. ELM server output provides links to the ELM annotation as well as to a number of remote resources. Using the links, researchers can explore the motifs, proteins, complex structures and associated literature to evaluate whether candidate motifs might be worth experimental investigation.
INTRODUCTION
Linear motifs (LMs) are short elements embedded within larger protein sequence segments that operate as sites of regulation (1–5). They can be found in telomeric proteins (6), in proteins of the extracellular matrix (7)—and seemingly every macromolecular complex in between. Many are post-translationally modified, but not all. The essence of their function is embodied in the linear amino acid sequence and is not dependent on the tertiary structural context. Nevertheless, as a consequence of low affinity binary binding interactions, they usually act in a concerted and cooperative manner, enabling regulatory decisions to be made on the basis of multiple inputs (8–12). These properties may be important for the inherent robustness of cellular systems (13), as cell regulation is increasingly revealed to be cooperative, networked and redundant in nature (14–20).
Over the time that we have worked to develop the Eukaryotic Linear Motif resource ELM, our conviction has grown that there will be well over a million LM instances in a higher eukaryotic proteome. (Phosphoproteomics is on the way to revealing ≫100 000 phosphorylation sites, for example.) If these estimates reflect reality, one might expect that experimentalists should be stumbling across new motifs with every experiment. But they are not. The paradox is that it remains difficult to establish the existence of LM instances whether by experiment or computationally. The bioinformatics problem is simple to state: LMs are too short (and the information content too poor) to be statistically significant in protein sequence searches. Experimentalists are similarly afflicted: while trying to identify LMs, they are likely to spend a lot of resources, time and effort performing experiments on the false motif candidates, which usually vastly outnumber the genuine ones in any set of proteins of interest (1).
Nevertheless, useful advances are now being made in the bioinformatics tools that address the remarkable modularity of eukaryotic regulatory proteins. Thus, two dedicated LM databases now exist: ELM (21) and the Minimotif Miner (22). (Users should utilize both resources as there are many differences in approach and the datasets only partially overlap.) Specialized databases for phosphorylation sites include PhosphoSite, Phospho.ELM and Phosida (23–25). Resources such as HPRD (26) and UniProtKB/Swiss-Prot (27) annotate a broader range of Post-Translational Modifications (PTMs). Furthermore, numerous predictive tools for identifying natively disordered protein segments—the main harbour for LMs (28–30)—have become available (31,32), complementing the more established globular domain resources Pfam, SMART, PROSITE and InterPro (33–36). The ELM datasets have been used by bioinformaticians to develop and benchmark novel prediction strategies such as hunting for motifs in interaction data and to provide likelihood estimates for motif candidates based on structural and sequence conservation contexts (37–41). While LM discovery remains challenging, if progress continues apace, it should become possible to address the intricate subfunctionalization of proteins like p53, CBP/p300, APC and Tau with ever-greater effectiveness.
Here, we provide an overview of the current status of the ELM resource and the research contexts in which it is being used. The utility of ELM is threefold: for researchers, it is first a knowledgebase, second a predictive tool but ELM has a third important function too; it can also be used for more general educational purposes, as it covers a topic that is often poorly served in text books. ELM provides written text summaries and links to the experimental literature that are a useful starting point for people who, for any reason, wish to gain an understanding of the role of LMs in cell regulation. We also take the opportunity here to provide a summary of progress made by the pioneering community of bioinformatics teams that are applying ELM to develop new tools for LM discovery. Finally, we provide some guidance about good practice and warnings about pitfalls for researchers seeking to apply ELM in experimental motif discovery.
WHAT ARE LMs?
To use ELM effectively, a user will need to grasp why such a resource is needed. The earliest definition of LM known to us was written in 1990 by Tim Hunt to introduce the new Protein Sequence Motifs column in Trends in Biological Sciences (42).
The sequences of many proteins contain short, conserved motifs that are involved in recognition and targeting activities, often separate from other functional properties of the molecule in which they occur. These motifs are linear, in the sense that three-dimensional organization is not required to bring distant segments of the molecule together to make the recognizable unit. The conservation of these motifs varies: some are highly conserved while others, for example, allow substitutions that retain only a certain pattern of charge across the motif.
This definition was written at a time when it was becoming apparent that many cellular proteins would have complex multidomain architectures and the first LMs such as KDEL, NLS, the Destruction Box of cyclin B and the fascinating KFERQ starvation-dependent lysosomal targeting motif were being reported (43–46). The definition has stood the test of time and can still serve very well today.
Sequence motifs contributing to the tertiary structure and primary function of globular domains are excluded by the definition of LM. An LM is effectively an irreducible unit of structure and function. Although LMs may be found in exposed parts of globular folds, they must be able to function independently to fit the definition: conversely, the globular domain would still have the same function if the LM was inactivated, although of course that domain function might well be dysregulated in the absence of the motif. The need to separate motif/domain functions applies to methods that seek to define new motifs. Historically, it has been difficult to develop computational methods that can distinguish short conserved segments of protein domains from LMs. Failure to make the distinction is likely to lead to false LM assignment (1), as has often happened for the nuclear export sequence (NES) as discussed by Hantschel et al. and Kadlec et al. (47,48).
Over the last few years, it has become increasingly clear that most LMs do not reside inside globular domains but instead are present in segments of natively disordered polypeptide. Often many LMs are clustered within one segment of native disorder. LMs quite frequently overlap, providing the potential for switch-like mutually exclusive functionality. For example, overlapping peptides from p53 are present in solved structures of several different protein complexes (20). Therefore, an overview of the types and locations of protein architecture modules existing in regulatory proteins provides an essential adjunct to LM investigation.
ELM RESOURCE ARCHITECTURE
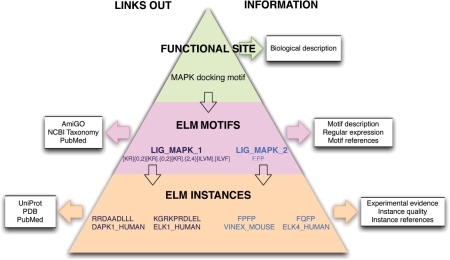
At the core of the ELM resource is a PostgreSQL relational database with 69 tables storing data about LMs. Not all of this complexity is fully utilized: it anticipates current and future filtering strategies as well as information retrieval by users. The key information content is summarized in Figure 1. Users should make sure they grasp the importance of the three fundamental nodes in the hierarchy: the top level ‘Functional Site’ links to ‘ELM Motif’ which includes ‘ELM Instances’. The top level of ‘Functional Site’ is essentially a biological designation with general information: for example, ‘Nuclear export signal’. The ‘ELM Motif’ is given a more specific description, links to information pertaining to the given LM, including key literature and Gene Ontology (GO) assignments, and includes the Regular Expression pattern representing the motif: see, for example, the NES entry at http://elm.eu.org/elmPages/TRG_NES_CRM1_1.html. Of note, ELM is effectively motif-centric—if a regular expression cannot be defined, there is no entry in ELM. An ‘ELM Instance’ embodies the specific information for a motif match in a protein sequence: for example, click on the links for the NES instance in MAPKAPK2. The instances provide the essential information that supports the ELM hierarchy. Instance-containing sequences are mapped to their respective UniProt entries. A well-annotated instance may also have links to the experimental literature, the types of experiments undertaken and to informative structure entries in the PDB (49). Importantly, an instance may have a reliability value assigned by the curator: many false positive motifs have been claimed in the literature. (Note: some of the older ELM entries do not yet have well-annotated instances).
Figure 1.
The ELM Resource hierarchy represented as a pyramid. ‘Functional Site’ provides a general description of the biology, for example, MAP Kinases have a docking motif in their substrates. There are more than one class of MAPK docking motifs and ELM currently provides two ‘ELM Motif’ entries. These contain the motif regular expression and are annotated with more specific information as well as linking out to remote resources including PubMed, NCBI Taxonomy and GO. At the base of the pyramid are the ‘ELM Instances’ that belong to a given ‘ELM Motif’ entry. The instances are annotated with information about experimental methods and instance quality and link to external resources including UniProt, PDB and PubMed.
All data input is by manual curation. Annotating each ELM entry typically involves extensive literature searches, BLAST runs, multiple alignment of relevant protein families, perusal of Swiss-Prot and other online databases and, where practical, discussion with experimentalist experts from the field. In order to promote interoperability with other bioinformatics resources, we use two public annotation standards. GO identifiers are used for cell compartment, molecular function and biological process (50) while the NCBI taxonomy database identifiers (51) are used for taxonomic nodes at the apex of phylogenetic groupings in which an LM occurs. A third standard—POSIX regular expressions (http://standards.ieee.org/regauth/posix/)—is used to represent the motif patterns. These ‘RegExps’ are conveniently usable in the Python and Perl scripting languages. They are analogous to PROSITE motifs (35), but with a different syntax. For example, the C-terminal motif LIG_CAP-Gly_1 that binds to CAP-Gly domains for microtubule plus-end regulation (52) is represented by the RegExp
[ed].{0,2}[ed].{0,2}[edq].{0,1}[YF]$
where $ is the protein C-terminus, preceded by a conserved aromatic residue and a flexibly spaced run of negatively charged residues. See the help page http://elm.eu.org/help.html#regular_expressions for guidance on the ELM expressions.
Table 1 provides some representative examples of different motif categories. Based on the type of function of the LM, we have defined four classes of ELM motif (Cleavage, Ligand, Modification and Target), which are summarized in the table. Some of these motifs have complicated regular expressions, others are very simple, e.g. with just two conserved positions. It has become clear that the most common conservation pattern is for three (semi-) conserved positions in the motif. A substantial minority of motifs have one or more positions that tolerate gaps (indels). The length range of indels can usually be accurately determined from sequence alignments: the most common indel is to allow a one-residue insertion.
Table 1.
The four classes of LM in the ELM classification and some representative examples
| Class | Class description | ELM_ID | Regular expressiona | ELM description |
|---|---|---|---|---|
| LIG | Motifs acting as ligands to globular protein domains. | LIG_MAPK1_1 | [KR]{0,2}[KR].{0,2}[KR].{2,4}[ILVM].[ILVF] | MAPK interacting molecules (e.g. MAPKKs, substrates, phosphatases) carry docking motifs that help to regulate specific interactions in the MAPK signalling networks. The classic motif approximates (R/K)xxxx#x# where # is a hydrophobic residue. |
| LIG_APCC_Dbox_1 | .R. .L. .[LIVM]. | An RxxL-based motif that binds to the Cdh1 and Cdc20 components of APC/C thereby targeting the protein for destruction in a cell cycle dependent manner. | ||
| TRG | Motifs within proteins that are sufficient for recognition and targeting to subcellular compartments. | TRG_AP2beta_CARGO_1 | [DE].{1,2}F[^P][^P][FL][^P][^P][^P]R | AP-2 beta appendage platform subdomain (top surface) binding motif used in targeting cargo for internalization. |
| TRG_PEX_1 | W … [FY] | Specific ELM present in Pex5p and binding to Pex13p and Pex14p. Part of the peroxisomal matrix protein import system | ||
| MOD | Sites of post-translational modification of proteins. | MOD_N-GLC_1 | .(N)[^P][ST]. . | Generic motif for N-glycosylation at Asparagine residues. Extracellular proteins are glycosylated in the Endoplasmatic Reticulum. The first step of the process, attachment of the carbohydrate precursor, is coupled to translation and import of the nascent polypeptide, preceding folding of the protein. |
| MOD_ProDKin_1 | …([ST])P. . | Proline-Directed Kinase (e.g. MAPK) phosphorylation site in higher eukaryotes. | ||
| CLV | Cleavage sites recognized by proteases for the processing of precursor proteins into biologically active products. | CLV_TASPASE1 | Q[MLVI]DG. .[DE] | Taspase1 is a threonine aspartase which was first identified as the protease responsible for processing the trithorax (MLL) type of histone methyltransferases. |
| CLV_PCSK_FUR_1 | R.[RK]R. | Furin (PACE) cleavage site (Arg-Xaa-[Arg/Lys]-Arg-|-Xaa) |
aRegular expression help is available at: http://elm.eu.org/help.html#regular_expressions.
Table 2 provides a summary of the data that have so far been entered into the ELM DB in its current state. The most noteworthy numbers are 146 ELM motifs, the >1300 instances and the >1100 citations of LM literature. Our goal is to create representative, not comprehensive, LM entries. For abundant motifs like the sumoylation site, with thousands of instances per proteome, we will not try to annotate more than a small fraction of experimental instances, since the appropriate location for these data are the protein annotation resources such as Swiss-Prot and HPRD.
Table 2.
Summary of the data stored in the ELM RDB
| Number of functional site entries | ELM motifs | Instances | Links to PDB structure entries | Go terms | PubMed links | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Totals | 110 | 146 | 1327 | 100 | 308 | 1125 | ||||
| By category | LIG | 89 | Human | 828 | Biological process | 152 | From ELM motif | 704 | ||
| MOD | 30 | Mouse | 104 | |||||||
| TRG | 19 | Rat | 65 | Cell compartment | 69 | From instance | 683 | |||
| CLV | 8 | Fly | 47 | |||||||
| Yeast | 88 | Molecular function | 87 | |||||||
| Other | 195 | |||||||||
ELM is primarily developed and deployed with open source software and is hosted on CentOS Linux. Pipeline software is mainly developed in Python including some modules from the http://BioPython.org project to retrieve information from SWISS-PROT and PubMed. The web interface software uses the CGImodel framework (53). The server output is HTML and Javascript.
WHY USE REGULAR EXPRESSIONS IN ELM?
The three most commonly used methods for bioinformatical representation of sequence conservation patterns are: Profile/HMMs (54); Artificial neural networks (ANNs) (55); and RegExps (http://en.wikipedia.org/wiki/Regular_expression). Of these, RegExps are considered the worst approach to encapture protein sequence information. They are ad hoc—typically created by annotators without applying a consistent formalism. The motif characters are represented with integer values, so RegExps cannot use position-weighting to capture weaker preferences. They are over-determined and can only capture exactly what is specified (whereas the more probabilistic HMMs and ANNs can rank near misses too). They do not support searching for an exact number of a given amino acid character within a specified range [which would better approximate the charged runs in e.g. CAP-Gly and NLS motifs (56)]. Despite these shortcomings, using RegExps to establish ELM has proved to be the correct decision. Many LMs have short indels in the pattern. HMM software does not (yet) provide for variable gaps with exactly bounded ranges while ANNs do not account for gaps at all: a motif such as the NES with multiple short indels is hard to represent with these algorithms. The scoring of presence/absence matches for LM RegExps simplifies statistical analyses of motif searches. These two advantages have been critical to the first wave of development of motif-hunting software.
Thus we consider that it was appropriate to initiate LM database resources with RegExps. Of course, HMMs and ANNs are used in a number of useful predictive tools, e.g. Scansite (57) and NetPhorest (58) and there is little doubt that HMMs, neural networks and other methods will grow in importance for LM analyses in future, once the contexts can be better controlled.
ACCESSING ELM
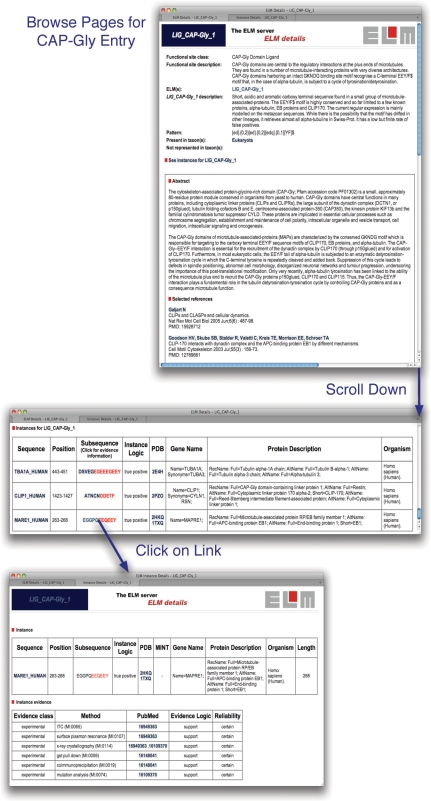
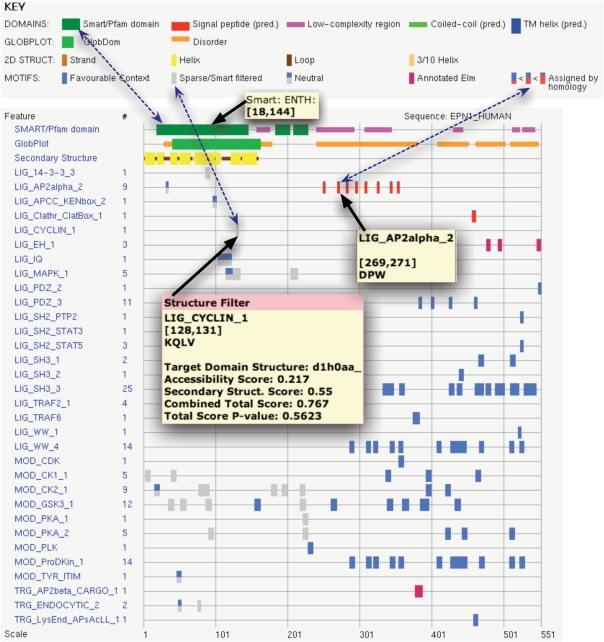
The ELM resource is freely accessible to users. The data in ELM can be accessed via the Web either interactively or programmatically. Motif entries are available to be browsed from the browse links page at http://elm.eu.org/. Details from the browse page for the LIG_CAP-Gly_1 entry are shown in Figure 2. A user can also submit a protein sequence of interest through the main submission page and will receive an output page with the matched candidates. The key data retrieved by the ELM resource for the sequence is displayed in a ‘bar code’ style graphical output as shown for the motif-rich endocytic protein Epsin-1 (Figure 3). Mouse-over provides annotation and there are many links to summaries in tabular and text form. Help is available online to explain the meanings of the elements and colour code in the output.
Figure 2.
Details from browse pages for the entry LIG_CAP-Gly_1 (http://elm.eu.org/elmPages/LIG_CAP-Gly_1.html). The upper window shows the description and the regular expression for the motif. Scrolling down past the references and the GO terms (not shown) leads to the table of known instances (middle window). Key information in the table includes whether an instance is a true positive, a link to the UniProt sequence entry and, if available, links to PDB structure entries (49). Clicking on the linked sequence for the instance in the EB1 protein (MARE1_HUMAN) opens a new page summarizing the annotated experimental evidence for the given instance. In this case, the motif has been exhaustively analysed and the supporting evidence is solid.
Figure 3.
Graphic from the output page of the ELM server queried with Epsin-1 sequence from the UniProt entry EPN1_HUMAN. The key indicates the content of the various coloured bars, e.g. the three connected by dotted arrows. Thirteen true LM instances are annotated either in this sequence or an orthologue from another species (magenta and red bar codes, respectively). Mouseover provides panels with different information depending on context, three examples of which are shown. One indicates an ENTH domain retrieved from SMART. A second points at an annotated DPW motif. The third mouseover provides the most detail: a structure for the ENTH domain (PDB entry d1h0) was used by the SF (41) to report that a cyclin motif candidate is too buried to be significant. Clicking on any object in the graphic will link to further details.
Programmatic access takes advantage of SOAP/XML Web Services (WS) interfaces for six ELM resource modules listed in Table 3. [See the EMBRACE registry for a large collection of Bioinformatics WS (59)]. Programmers can use the ELM DB WS interfaces to collect data—for example, a query might be to retrieve all regular expressions stored in ELM or another query might be for all ELM instances, or a defined subset thereof. Other WS interfaces allow LM matching to a query sequence and structural and conservation filtering.
Table 3.
Web Service interfaces for the ELM tool suite
| Resource module | Purpose of resource module | Links to WSDLs |
|---|---|---|
| ELM Database | Retrieve data stored by ELM | http://elm.eu.org/webservice/ELMdb.wsdl |
| http://api.bioinfo.no/wsdl/ELMdb.wsdl | ||
| ELMMatcher | Map ELM Motifs to query sequence | http://elm.eu.org/webservice/wsELMMatcher.wsdl |
| http://api.bioinfo.no/wsdl/ELMMatcher.wsdl | ||
| ELM CS Filter | Evaluate conservation of LM matches in reference sequence | http://conscore.embl.de/webservice/CS.wsdl |
| ELM SF | Evaluate accessibility and structure context of LM matches in query sequence given a reference structure | http://structurefilter.embl.de/webservice/structureFilter.wsdl |
| GlobPlot | Evaluate disorder propensity in query sequence | http://globplot.embl.de/webservice/globplot.wsdl |
| Phospho.ELM | Retrieve phosphorylation data stored by Phospho.ELM | http://phospho.elm.eu.org/webservice/phosphoELMdb.wsdl |
Upon request, we can provide a SQL dump if for any reason, the WS interface is not suitable. At some future point, we would like to provide a standardized ELM DB dump, probably using the BioMart format (60).
THE ELM RESOURCE FILTERS
Searches of sequence databases with short motifs do not yield significant results (due to the large number of non-functional sequences matching the motif consensus) and therefore, it is necessary to evaluate the context of the match. Essentially, any aspect of a protein that can be informative might provide contextual filtering. Filters might be simple or complicated and ELM provides examples of both. Originally, three simple filters (21) were implemented in ELM: (i) Cell compartment filter: an LM is only meaningful in appropriate cell compartments; (ii) Taxonomy filter: an LM is only meaningful in an organism that is known to possess its interaction partners; and (iii) SMART globular domain filter: LMs are interaction sites and must be accessible, hence they are much more common in natively disordered sequence. ELM does not provide benchmarked scores for the simple filters. Two more complicated filters have been implemented and benchmarked to provide reliability assessments, for structural context and evolutionary conservation.
The ELM structure filter (SF) assesses the accessibility and secondary structure components of LM candidates whenever a reference globular domain structure is available (41). The benchmarked scale shows that most LMs are in exposed and accessible loops. Although a few genuine LMs are quite inaccessible in the available structural conformation, the benchmarking indicates that it is usually not worth experimental testing of the inaccessible motifs unless there is an indication of, for example, allosteric rearrangement that might enable the site to become exposed. When it applies, the SF is much more informative than the simple globular domain filter. The SF is implemented in the ELM resource output (Figure 3), and can be accessed independently as a web service (Table 3).
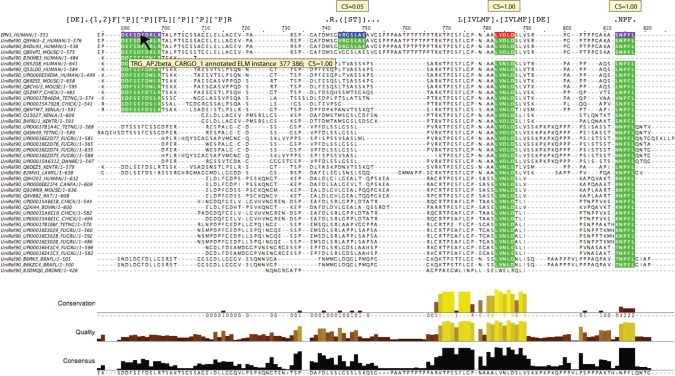
The ELM conservation score (CS) filter assesses the conservation of motif candidates in related proteins (61). LMs tend to be more evolutionarily dynamic than globular domains—it is uncommon to find an LM instance that is conserved between yeast and mammals (e.g. see the GLEBS and FFAT motif entries for counterexamples). The CS filter is a pipeline to collect and align homologous sequences and test ELM motifs for conservation, using a benchmarked scoring scheme. The CS filter has already proven its value in motif discovery efforts (62,63) but, due to the resource reengineering required, is not yet implemented in the ELM output. For the time being, therefore, it is offered as a stand-alone server (http://elm.eu.org/conscorer) and web service (Table 3). Figure 4 shows variation in conservation of some of the motif matches from the Epsin-1 example used above (Figure 3).
Figure 4.
Representative results from the CS web interface, displayed with the annotated sequence alignment editor JalView (86). The alignment shows the set of sequences obtained by the CS filter with the human Epsin1 query sequence at top: the sequences belong to several paralogous families of Epsins. Four motif matches are highlighted in the reference sequence (magenta, annotated in this sequence; red, assigned by the instance mapper; blue, unannotated match) and in other sequences that align to the reference motif (green). The left-most match is a known instance of TRG_AP2beta_CARGO_1 and gives a top score of 1.00 despite only being present in sequences belonging to two of the Epsin paralogues. This is because most sequences that lack the motif have gaps aligned to it that do not affect the CS score. The second motif is a candidate instance for MOD_PKA_2 but is poorly conserved, scoring 0.05. This candidate would probably not be worth investigating unless there was prior evidence of phosphorylation at the site. The remaining two motifs are known instances of LIG_Clathr_ClatBox_1 and LIG_EH_1, which obtain the maximum CS score since they are conserved in all Epsin paralogues.
THE ELM INSTANCE MAPPER
It is not uncommon that all the experimentation demonstrating the existence of a particular LM instance has been undertaken in a single model organism, e.g. yeast, or cell lines from one of mouse, chicken or human. For a given LM class, the set of known instances may have been identified in a range of different species. Therefore, researchers are routinely faced with the issue of mapping experimental results from diverse organisms onto the protein sequence of their model organism. The instance mapper module addresses this issue for the ELM server.
A rarely used BLAST variant, PHI-BLAST, is at the core of the ELM instance mapper (64). PHI-BLAST requires a regular expression in addition to the query sequence: the pattern must have at least one match in the query. We found PHI-BLAST to be ideally suited for mapping known LM matches from homologous sequences, so that the instance mapping issue was reduced to developing a protocol to utilize it effectively.
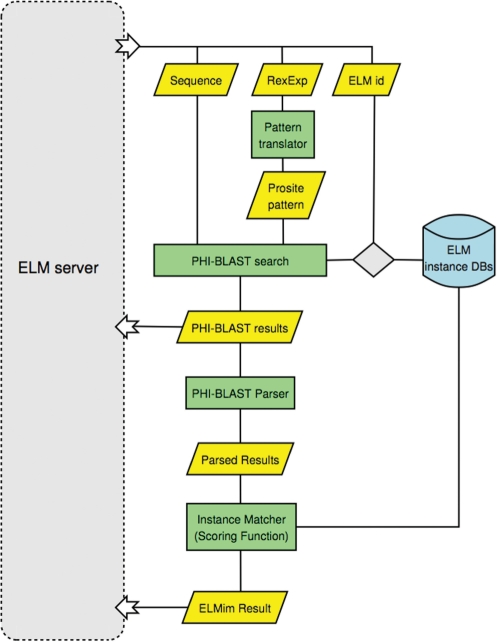
The flow scheme of the instance mapper is summarized in Figure 5. Sequences harbouring known instances are stored in a small BLAST formatted database. For each pattern matching the query, this database is searched by PHI-BLAST. The instance mapper then parses the output and assigns a divergence-based score to any matches that are retrieved. These are then displayed in the ELM server graphical output (Figure 3).
Figure 5.
Flow scheme for the ELM Instance Mapper. For each predicted LM from an ELM database search, a PHI-BLAST search is performed against a database containing all sequences with known instances of the predicted LM. Input to PHI-BLAST is the query sequence and the ELM Regular Expression (which is adapted for use with PHI-BLAST). Each of the aligned motifs, between query and ELM instance sequence, are evaluated and scored (see main text). If the motif in the ELM instance sequence is a known instance, and the calculated score is above a threshold (Sei ≥ 0.3), it is reported as a mapped instance. Both the ELM instance mapper and the underlying PHI-BLAST results are returned to the ELM server, for the user to inspect.
PHI-BLAST calculates an E-value, based on the BLAST bit score, which is useful for determining the statistical significance of a given alignment. However, this statistic does not reflect how similar the query sequence is to the LM instance sequence, which is particularly relevant for our purpose. To address this issue, we have devised an ELM instance score Sei that is calculated from the PHI-BLAST alignment:
where i is the number of identical positions in the alignment, g is the number of gaps, la is the length of the alignment (minus gaps), lq is the length of the query sequence and ls is the length of the subject sequence. The assumptions behind the score are that false matches are more likely at higher divergence and in longer sequences. At higher divergence, the sequences may be nonorthologous (or only partially so) or, in orthologous sequences, nonorthologous matches may also be superposed, especially for common, simple motifs. Therefore, while the instance matcher can retrieve genuine instances in sequences that are as low as 30% identity, a low score serves as a warning to evaluate the match. Note that this score is designed for evaluation of pairwise matches: if we had a multiple alignment and were confident that the alignment was correct for a motif, then the conservation can be scored as ‘more’ significant at higher divergence (61).
The instance mapper is a key addition to the resource as it unites the information content of the experimental instances stored in the ELM database with the motif exploration capabilities afforded by the ELM regular expressions.
USER COMMUNITY FEEDBACK AND INTERACTION
In common with other bioinformatics resources, only a few of the ELM users choose to communicate with us. Users should know that certain types of communication are very useful to us. Obviously, if a server problem persists for a few hours, we should be informed immediately. Suggestions about the ELM resource interface would also be welcome—though we can probably only respond slowly to good ideas.
Of most use to ELM and the user community would be information to improve the data stored in ELM. Sometimes this might be a simple update such as an important instance that has been omitted, a new structure or a useful reference. More substantial help with creating or improving entries would be particularly valuable. In several cases, experts have contributed or reviewed entries for ELM. Entries with expert involvement include: LIG_CAP-Gly_1, LIG_EH_1, LIG_SxIP_EBH_1, LIG_ULM_U2AF65_1, LIG_RRM_PRI_1, TRG_AP2beta_CARGO_1 (65–70).
The obvious reason why researchers may be chary of getting involved with improving ELM is the time and effort that it costs. There is an upside that scientific information now disseminates to a great extent through the web: ELM can provide another route to showcase your work and, presumably, the prouder you are of your achievements, the more visible you would like them to be. We thank those researchers who have already helped us improve ELM and hope that their research will receive some reciprocal benefit.
ROLE OF ELM IN LM RESEARCH/DISCOVERY
As ELM has become more widely known to researchers, experimental investigations of candidate matches to known motifs have begun to appear in the literature. For example, an HCMV transmembrane protein has been shown to have LMs for cooption of cellular retention systems, aiding viral immune evasion (71). A candidate 14-3-3-binding phosphosite has been validated in the cytosolic C-terminus of integrin-α4 (72). Several regulatory motifs have been investigated in Drosophila cryptochrome, a regulator of circadian rhythm (73). Collectively such studies afford optimism that our work to establish the ELM resource will increasingly be justified by experimental application.
We take the view that by applying ELM ourselves, we can better evaluate and optimize our methodologies. We have sometimes been able to employ a protocol involving GO term enrichment to reveal sets of proteins with LM matches that are significantly enriched in specific contexts. Thus, we have reported a bioinformatics survey (63) of KEN box anaphase destruction motifs enriched in mitotic proteins: KEN box motifs in CHFR and C13orf3 are thought to aid in defining their roles in mitosis, though experimental validation is still needed (74,75). In a second example, while annotating the SUMO motif, we were able to define a larger motif, KEPE, superposed on a subset of sumoylation sites (62). It is, however, too soon for the role of KEPE to have been investigated.
The ELM instance dataset has been deployed by several bioinformatics groups in ways that have provided insight into LM context and/or to develop and benchmark novel strategies for LM discovery. Thus, the anecdotal observation that LMs are more abundant in natively disordered protein sequence (21) has been verified by more systematic analyses using benchmarked native disorder predictors (28,29). More recently, this research line has been extended with the ANCHOR server providing benchmarked prediction of short stretches of sequence that have strong interacting potential (76). The local context of LMs has been further investigated, revealing that the adjacent peptide sequence often has a role in modulating LM function (77,78). Stemming from an awareness that viruses utlilize numerous LMs to hijack cellular systems, Dinkel and Sticht (37) developed and benchmarked a pipeline to apply conservation and domain masking to motif candidates. Observing that multiple sequence alignment software has been overtrained on globular sequences and therefore performs quite poorly with short conserved motifs, the BAliBASE alignment benchmark suite was extended with an LM benchmark in the hope that this will lead to improved alignment algorithms (79).
While the ELM resource per se is not suited to de novo discovery of hitherto unknown motifs, the instances have been used by others to develop and benchmark tools for just this purpose. Yeast 2-hybrid data includes candidate LM-mediated interactions and both DILIMOT and SLiMFinder use interaction sets to search for enriched motifs in the binders of a protein (38,39,80). These methods depend on overrepresentation of a motif and therefore are probably not suited to motifs that have few biological instances. However, another promising approach uses amino acid preferences to sample 3D structural surfaces for sites with high peptide binding values (40): such methods have the potential to reveal LMs that have only a single functional instance in a proteome. These strategies illustrate how other data (interactions, structures) can be integrated into bioinformatics LM discovery pipelines, complementing experimental approaches for motif definition such as peptide libraries and arrays (81–83).
When we began the ELM project, LM bioinformatics was essentially nonexistent (21). The progress in the last few years has been impressive and exciting. There is growing awareness that the study of protein interactions is not just about globular–globular interfaces (5,84). Protein interaction data and domain surfaces can now be explored for possible LM interactors. There is much more to be done before researchers can pull up strong LM candidates as easily as running BLAST searches, but this goal—so important if we are to understand cell regulation—no longer seems to be impossibly fanciful.
EVALUATING AND APPLYING THE ELM SERVER RESULTS
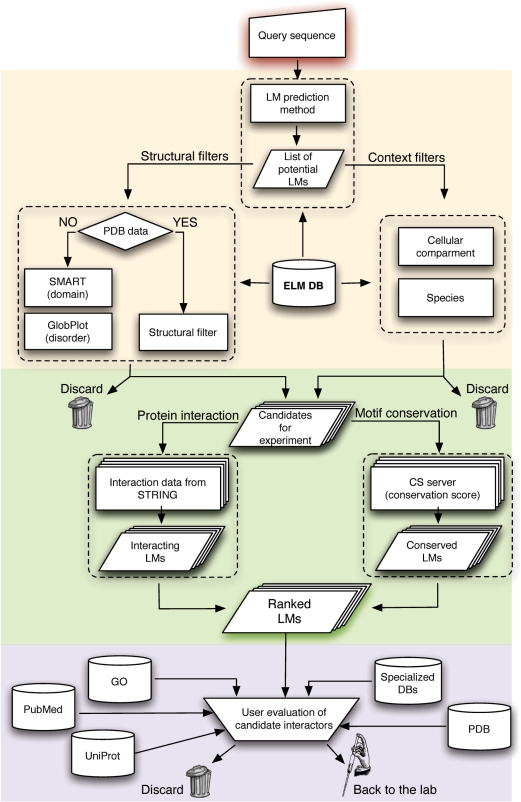
Candidate LMs require experimental validation. The key to using ELM is to select good candidates for experimental validation and not waste time on the poor ones. Since LMs are always interaction sites, they must be in the same cell compartment as their ligand. There is little point in experimentally testing a candidate cyclin-binding motif in a collagen sequence. Likewise, a motif that is deeply buried in a solved structure makes a poor choice for experimentation (41). Therefore, it is first necessary to establish if a motif match is conserved, exposed and in the right cell compartment, according to the ELM filters. Motifs that pass these tests can then be further examined using a range of bioinformatics tools. Figure 6 shows a flowchart for how a typical motif evaluation might proceed. After the initial ELM tests, native disorder predictors and domain databases can give an indication of structural context. If the motif is within a known 3D structure, the context should be visualized; e.g. with PyMol (http://pymol.sourceforge.net/). Swiss-Prot features, the HPRD entry and phosphorylation databases may provide additional structure–function context. A user should always prepare a multiple sequence alignment and examine the motif conservation. Note that multiple alignment software sometimes struggle with motif alignments, with MAFFT (85) perhaps being the best current choice (79). If motifs are present but misaligned, an alignment editor such as JalView (86) may be helpful. Is the motif conserved in a specific lineage, e.g. vertebrates? If the motif is conserved, is the adjacent sequence less so? If things are looking good, it is important to ask whether the proposed LM function makes any sense for the protein; if this is unfamiliar, it is advisable to spend some time reading the literature: the ELM links to PubMed are a useful starting point, but unlikely to be exhaustive.
Figure 6.
Workflow diagram illustrating how a user might explore LM candidates with ELM. The pipeline proceeds through three main phases utilizing the ELM resource (beige background) ELM associated tools (green) and more general bioinformatics resources (pink). Candidate LMs can be rejected by ELM filters if in unsuitable contexts. Sequence conservation and enrichment in interaction data using DiLiMot or SLiMFinder can provide additional scores to rank motifs. In the final phase any potentially relevant bioinformatics resources should be examined to provide further context to motif candidates. If promising candidates survive this process, the end point of the bioinformatics pipeline has been reached and laboratory validation is now required.
If LM candidates have survived the routine tests, there are other bioinformatics tools that might provide further insight. Protein interaction resources such as STRING (87), MINT (88) and IntAct (89) can reveal if a ligand protein is known to be close in the network. Interaction data can also be supplied to DILIMOT and/or SLiMFinder to evaluate whether there is statistical support for motif enrichment (38,39). Enrichment of motifs with UniProt GO terms and other keywords can sometimes provide statistical support for sets of motifs (62,63,90). SIRW is an online tool (http://sirw.embl.de/index.html) that allows keyword exploration for RegExps (91). If enrichment is found, SIRW can provide a probability estimate using Fisher’s Exact Test. Of course, motif enrichment can be an artefact of sequence length or amino acid bias so judgement of the results is required. If the enriched set is not more conserved than the background, then it is unlikely to be biologically meaningful.
After doing all this, ask once again: Is the motif buried? We think it likely that inaccessible motifs are the most common reason for erroneous LM reports in the literature.
Even when an LM candidate is in the right cell compartment, and survives many other tests, it does not have to be functional as it still may never contact the ligand protein (20). There is increasing evidence that cell signalling decisions are made in large dynamic protein complexes. If a motif-containing protein is never in the same complex as a ligand protein, the motif will be false. For this reason, cell localization assays are useful, although they can be misleading if overexpression is used. Coimmunoprecipitation and pull down experiments are also widely used as part of motif validation. We thought it might be of interest to list the most commonly annotated methods applied in motif validation and these are presented in Table 4. Since no one experiment is definitive, many of these methods will have been applied to a well-validated motif instance.
Table 4.
The main experimental methods used in motif validation, as recorded in ELM
| Experimental method | PSI-MI IDa | Number of occurrences |
|---|---|---|
| Mutation analysis | MI:0074 | 305 |
| Pull down assay | MI:0096 | 200 |
| Yeast 2 hybrid assay | MI:0018 | 115 |
| Co-immunoprecipitation | MI:0019 | 98 |
| X-ray crystallography | MI:0114 | 75 |
| Motif Deletion | MI:0573 | 53 |
| Competitive binding assay | MI:0405 | 39 |
| Protein overlay assay | MI:0049 | 38 |
| Colocalization by immunostaining | MI:0022 | 37 |
| Nuclear magnetic resonance | MI:0077 | 30 |
| Isothermal titration calorimetry (ITC) | MI:0065 | 29 |
| Protein truncation mutants | 28 | |
| Immunological detection and localization | MI:0422 | 27 |
| Mass spectrometry | MI:0427 | 24 |
| Motif transplantation | 20 | |
| Western blot | MI:0113 | 19 |
| Radiolabelling/pulse chase | MI:0517 | 19 |
| Surface plasmon resonance | MI:0107 | 15 |
aIdentifier for the HUPO PSI-MI exchange standard entry that either defines or encompasses the listed experiment (92).
CURRENT LIMITATIONS AND FUTURE DIRECTIONS
In common with LM bioinformatics, in general, ELM has advanced to a state of practical usefulness, yet there is much more to do. LM RegExp matches cannot yet be taken as indicators of true functional sites and the candidates must be experimentally verified. The ELM dataset is incomplete with respect to motifs reported in the literature and there is work to be done to extend the coverage of the database: currently, users should not use ELM as a sole source of LM information. We have identified a need to improve the data captured regarding interactions of the ELM instances, which currently are of limited use for systems modelling in silico. ELM filtering can be improved in the short to medium term by embedding the CS filter and by using Swiss-Prot topology domains for automated cell compartment filtering of transmembrane proteins. In the ELM output, we would like to present the user with phosphorylation sites and other readily available information about the structure/function modules of query proteins. It is our hope that most of these goals will have been achieved when we next report on ELM.
FUNDING
The ELM Web Service interfaces were developed in the framework of the EU FP5 EMBRACE grant (LHSG-CT-2004-512092). The FIRB 2004 ITALBIONET grant (to A.V.); the NGFN DiGToP grant (to M.S.); the FP6 ProteomeBinders grant (to N.H.). SF development was aided by DAAD and Vigoni covered travel expenses between Heidelberg and Rome. Funding for open access charge: EMBL.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank the former contributors to the ELM resource, the Bioinformatics developers who have applied the ELM instances to develop discovery methods and the ELM resource users whose web access statistics spurred us on.
REFERENCES
- 1.Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 2.Neduva V, Russell RB. Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 2006;17:465–471. doi: 10.1016/j.copbio.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Kadaveru K, Vyas J, Schiller MR. Viral infection and human disease—insights from minimotifs. Front. Biosci. 2008;13:6455–6471. doi: 10.2741/3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox-Erlich S, Schiller MR, Gryk MR. Structural conservation of a short, functional, peptide-sequence motif. Front. Biosci. 2009;14:1143–1151. doi: 10.2741/3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petsalaki E, Russell RB. Peptide-mediated interactions in biological systems: new discoveries and applications. Curr. Opin. Biotechnol. 2008;19:344–350. doi: 10.1016/j.copbio.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 7.Salsmann A, Schaffner-Reckinger E, Kieffer N. RGD, the Rho'd; to cell spreading. Eur. J. Cell Biol. 2006;85:249–254. doi: 10.1016/j.ejcb.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 9.Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc. Natl Acad. Sci. USA. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright PE, Dyson HJ. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer BJ, Blinov ML, Loew LM. Molecular machines or pleiomorphic ensembles: signaling complexes revisited. J. Biol. 2009;8:81. doi: 10.1186/jbiol185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein A, Pache RA, Bernado P, Pons M, Aloy P. Dynamic interactions of proteins in complex networks: a more structured view. FEBS J. 2009;276:5390–5405. doi: 10.1111/j.1742-4658.2009.07251.x. [DOI] [PubMed] [Google Scholar]
- 13.Kitano H. Towards a theory of biological robustness. Mol. Syst. Biol. 2007;3:137. doi: 10.1038/msb4100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawson T, Kofler M. Kinome signaling through regulated protein-protein interactions in normal and cancer cells. Curr. Opin. Cell Biol. 2009;21:147–153. doi: 10.1016/j.ceb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volonte C, D'A;mbrosi N, Amadio S. Protein cooperation: from neurons to networks. Prog. Neurobiol. 2008;86:61–71. doi: 10.1016/j.pneurobio.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Whitty A. Cooperativity and biological complexity. Nat. Chem. Biol. 2008;4:435–439. doi: 10.1038/nchembio0808-435. [DOI] [PubMed] [Google Scholar]
- 18.Williamson JR. Cooperativity in macromolecular assembly. Nat. Chem. Biol. 2008;4:458–465. doi: 10.1038/nchembio.102. [DOI] [PubMed] [Google Scholar]
- 19.Tan CS, Bodenmiller B, Pasculescu A, Jovanovic M, Hengartner MO, Jorgensen C, Bader GD, Aebersold R, Pawson T, Linding R. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci. Signal. 2009;2:ra39. doi: 10.1126/scisignal.2000316. [DOI] [PubMed] [Google Scholar]
- 20.Gibson TJ. Cell regulation: determined to signal discrete cooperation. Trends Biochem. Sci. 2009;34:471–482. doi: 10.1016/j.tibs.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Puntervoll P, Linding R, Gemünd C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, et al. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajasekaran S, Balla S, Gradie P, Gryk MR, Kadaveru K, Kundeti V, Maciejewski MW, Mi T, Rubino N, Vyas J, et al. Minimotif miner 2nd release: a database and web system for motif search. Nucleic Acids Res. 2009;37:D185–D190. doi: 10.1093/nar/gkn865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 24.Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: a database of phosphorylation sites—update 2008. Nucleic Acids Res. 2008;36:D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.UniProt Consortium. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 29.Ren S, Uversky VN, Chen Z, Dunker AK, Obradovic Z. Short Linear Motifs recognized by SH2, SH3 and Ser/Thr Kinase domains are conserved in disordered protein regions. BMC Genomics. 2008;9(Suppl. 2):S26. doi: 10.1186/1471-2164-9-S2-S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell RB, Gibson TJ. A careful disorderliness in the proteome: sites for interaction and targets for future therapies. FEBS Lett. 2008;582:1271–1275. doi: 10.1016/j.febslet.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 31.Bourhis JM, Canard B, Longhi S. Predicting protein disorder and induced folding: from theoretical principles to practical applications. Curr. Protein Pept. Sci. 2007;8:135–149. doi: 10.2174/138920307780363451. [DOI] [PubMed] [Google Scholar]
- 32.He B, Wang K, Liu Y, Xue B, Uversky VN, Dunker AK. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 33.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dinkel H, Sticht H. A computational strategy for the prediction of functional linear peptide motifs in proteins. Bioinformatics. 2007;23:3297–3303. doi: 10.1093/bioinformatics/btm524. [DOI] [PubMed] [Google Scholar]
- 38.Edwards RJ, Davey NE, Shields DC. SLiMFinder: a probabilistic method for identifying over-represented, convergently evolved, short linear motifs in proteins. PLoS ONE. 2007;2:e967. doi: 10.1371/journal.pone.0000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neduva V, Russell RB. DILIMOT: discovery of linear motifs in proteins. Nucleic Acids Res. 2006;34:W350–W355. doi: 10.1093/nar/gkl159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petsalaki E, Stark A, Garcia-Urdiales E, Russell RB. Accurate prediction of peptide binding sites on protein surfaces. PLoS Comput. Biol. 2009;5:e1000335. doi: 10.1371/journal.pcbi.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Via A, Gould CM, Gemünd C, Gibson TJ, Helmer-Citterich M. A structure filter for the Eukaryotic Linear Motif Resource. BMC Bioinformatics. 2009;10:351. doi: 10.1186/1471-2105-10-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunt T. Protein sequence motifs involved in recognition and targeting: a new series. Trends Biochem. Sci. 1990;15:305. [Google Scholar]
- 43.Pelham HR. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 1990;15:483–486. doi: 10.1016/0968-0004(90)90303-s. [DOI] [PubMed] [Google Scholar]
- 44.Dingwall C, Laskey RA. Nuclear targeting sequences – a consensus? Trends Biochem. Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 45.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 46.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 47.Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, Superti-Furga G. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 48.Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat. Struct. Mol. Biol. 2004;11:330–337. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- 49.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gene Ontology Consortium. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinmetz MO, Akhmanova A. Capturing protein tails by CAP-Gly domains. Trends Biochem. Sci. 2008;33:535–545. doi: 10.1016/j.tibs.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 53.Chenna R, Gemünd C. cgimodel: CGI programming made easy with Python. Linux J. 2000;75:142–149. [Google Scholar]
- 54.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 55.Krogh A. What are artificial neural networks? Nat. Biotechnol. 2008;26:195–197. doi: 10.1038/nbt1386. [DOI] [PubMed] [Google Scholar]
- 56.Seiler M, Mehrle A, Poustka A, Wiemann S. The 3of5 web application for complex and comprehensive pattern matching in protein sequences. BMC Bioinformatics. 2006;7:144. doi: 10.1186/1471-2105-7-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, et al. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettifer S, Thorne D, McDermott P, Attwood T, Baran J, Bryne JC, Hupponen T, Mowbray D, Vriend G. An active registry for bioinformatics web services. Bioinformatics. 2009;25:2090–2091. doi: 10.1093/bioinformatics/btp329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart—biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chica C, Labarga A, Gould CM, Lopez R, Gibson TJ. A tree-based conservation scoring method for short linear motifs in multiple alignments of protein sequences. BMC Bioinformatics. 2008;9:229. doi: 10.1186/1471-2105-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diella F, Chabanis S, Luck K, Chica C, Ramu C, Nerlov C, Gibson TJ. KEPE—a motif frequently superimposed on sumoylation sites in metazoan chromatin proteins and transcription factors. Bioinformatics. 2009;25:1–5. doi: 10.1093/bioinformatics/btn594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Michael S, Trave G, Ramu C, Chica C, Gibson TJ. Discovery of candidate KEN-box motifs using cell cycle keyword enrichment combined with native disorder prediction and motif conservation. Bioinformatics. 2008;24:453–457. doi: 10.1093/bioinformatics/btm624. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Z, Schaffer AA, Miller W, Madden TL, Lipman DJ, Koonin EV, Altschul SF. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998;26:3986–3990. doi: 10.1093/nar/26.17.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weisbrich A, Honnappa S, Jaussi R, Okhrimenko O, Frey D, Jelesarov I, Akhmanova A, Steinmetz MO. Structure-function relationship of CAP-Gly domains. Nat. Struct. Mol. Biol. 2007;14:959–967. doi: 10.1038/nsmb1291. [DOI] [PubMed] [Google Scholar]
- 66.Rumpf J, Simon B, Jung N, Maritzen T, Haucke V, Sattler M, Groemping Y. Structure of the Eps15-stonin2 complex provides a molecular explanation for EH-domain ligand specificity. EMBO J. 2008;27:558–569. doi: 10.1038/sj.emboj.7601980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 68.Corsini L, Bonnal S, Basquin J, Hothorn M, Scheffzek K, Valcarcel J, Sattler M. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 2007;14:620–629. doi: 10.1038/nsmb1260. [DOI] [PubMed] [Google Scholar]
- 69.Rideau AP, Gooding C, Simpson PJ, Monie TP, Lorenz M, Huttelmaier S, Singer RH, Matthews S, Curry S, Smith CW. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat. Struct. Mol. Biol. 2006;13:839–848. doi: 10.1038/nsmb1137. [DOI] [PubMed] [Google Scholar]
- 70.Edeling MA, Mishra SK, Keyel PA, Steinhauser AL, Collins BM, Roth R, Heuser JE, Owen DJ, Traub LM. Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev. Cell. 2006;10:329–342. doi: 10.1016/j.devcel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 71.Maffei M, Ghiotto F, Occhino M, Bono M, De Santanna A, Battini L, Gusella GL, Fais F, Bruno S, Ciccone E. Human cytomegalovirus regulates surface expression of the viral protein UL18 by means of two motifs present in the cytoplasmic tail. J. Immunol. 2008;180:969–979. doi: 10.4049/jimmunol.180.2.969. [DOI] [PubMed] [Google Scholar]
- 72.Deakin NO, Bass MD, Warwood S, Schoelermann J, Mostafavi-Pour Z, Knight D, Ballestrem C, Humphries MJ. An integrin-{alpha}4-14-3-3{zeta}-paxillin ternary complex mediates localised Cdc42 activity and accelerates cell migration. J. Cell Sci. 2009;122:1654–1664. doi: 10.1242/jcs.049130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hemsley MJ, Mazzotta GM, Mason M, Dissel S, Toppo S, Pagano MA, Sandrelli F, Meggio F, Rosato E, Costa R, et al. Linear motifs in the C-terminus of D. melanogaster cryptochrome. Biochem. Biophys. Res. Commun. 2007;355:531–537. doi: 10.1016/j.bbrc.2007.01.189. [DOI] [PubMed] [Google Scholar]
- 74.Privette LM, Weier JF, Nguyen HN, Yu X, Petty EM. Loss of CHFR in human mammary epithelial cells causes genomic instability by disrupting the mitotic spindle assembly checkpoint. Neoplasia. 2008;10:643–652. doi: 10.1593/neo.08176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Theis M, Slabicki M, Junqueira M, Paszkowski-Rogacz M, Sontheimer J, Kittler R, Heninger AK, Glatter T, Kruusmaa K, Poser I, et al. Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. EMBO J. 2009;28:1453–1465. doi: 10.1038/emboj.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meszaros B, Simon I, Dosztanyi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stein A, Aloy P. Contextual specificity in peptide-mediated protein interactions. PLoS ONE. 2008;3:e2524. doi: 10.1371/journal.pone.0002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chica C, Diella F, Gibson TJ. Evidence for the concerted evolution between short linear protein motifs and their flanking regions. PLoS ONE. 2009;4:e6052. doi: 10.1371/journal.pone.0006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perrodou E, Chica C, Poch O, Gibson TJ, Thompson JD. A new protein linear motif benchmark for multiple sequence alignment software. BMC Bioinformatics. 2008;9:213. doi: 10.1186/1471-2105-9-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Neduva V, Linding R, Su-Angrand I, Stark A, de Masi F, Gibson TJ, Lewis J, Serrano L, Russell RB. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ferraro E, Via A, Ausiello G, Helmer-Citterich M. A neural strategy for the inference of SH3 domain-peptide interaction specificity. BMC Bioinformatics. 2005;6(Suppl. 4):S13. doi: 10.1186/1471-2105-6-S4-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Machida K, Thompson CM, Dierck K, Jablonowski K, Karkkainen S, Liu B, Zhang H, Nash PD, Newman DK, Nollau P, et al. High-throughput phosphotyrosine profiling using SH2 domains. Mol. Cell. 2007;26:899–915. doi: 10.1016/j.molcel.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 83.Zhu G, Fujii K, Liu Y, Codrea V, Herrero J, Shaw S. A single pair of acidic residues in the kinase major groove mediates strong substrate preference for P-2 or P-5 arginine in the AGC, CAMK, and STE kinase families. J. Biol. Chem. 2005;280:36372–36379. doi: 10.1074/jbc.M505031200. [DOI] [PubMed] [Google Scholar]
- 84.Stein A, Panjkovich A, Aloy P. 3did Update: domain-domain and peptide-mediated interactions of known 3D structure. Nucleic Acids Res. 2009;37:D300–D304. doi: 10.1093/nar/gkn690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 86.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8 – a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Copley RR. The EH1 motif in metazoan transcription factors. BMC Genomics. 2005;6:169. doi: 10.1186/1471-2164-6-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramu C. SIRW: A web server for the Simple Indexing and Retrieval System that combines sequence motif searches with keyword searches. Nucleic Acids Res. 2003;31:3771–3774. doi: 10.1093/nar/gkg546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C, et al. The HUPO PSI's; molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]