Abstract
The DNA Data Bank of Japan (DDBJ) (http://www.ddbj.nig.ac.jp) has collected and released 1 701 110 entries/1 116 138 614 bases between July 2008 and June 2009. A few highlighted data releases from DDBJ were the complete genome sequence of an endosymbiont within protist cells in the termite gut and Cap Analysis Gene Expression tags for human and mouse deposited from the Functional Annotation of the Mammalian cDNA consortium. In this period, we started a novel user announcement service using Really Simple Syndication (RSS) to deliver a list of data released from DDBJ on a daily basis. Comprehensive visualization of a DDBJ release data was attempted by using a word cloud program. Moreover, a new archive for sequencing data from next-generation sequencers, the ‘DDBJ Read Archive’ (DRA), was launched. Concurrently, for read data registered in DRA, a semi-automatic annotation tool called the ‘DDBJ Read Annotation Pipeline’ was released as a preliminary step. The pipeline consists of two parts: basic analysis for reference genome mapping and de novo assembly and high-level analysis of structural and functional annotations. These new services will aid users’ research and provide easier access to DDBJ databases.
INTRODUCTION
The DNA Data Bank of Japan (DDBJ) is one of three databanks that constitute the DDBJ/EMBL-Bank/GenBank International Nucleotide Sequence Database (INSD), which was established through close collaboration with the European Bioinformatics Institute (EBI) in Europe and the National Center for Biotechnology Information (NCBI) in the USA. DDBJ is administered by the Center for Information Biology and DDBJ (CIB-DDBJ) of the National Institute of Genetics (http://www.nig.ac.jp/index-e.html) with funding endorsement from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT). All researchers can submit their data to one of the three summit databanks to register it with INSD. The data that are enrolled are exchanged on every day, so that the three collaborating databanks share virtually the same data at any given time. The syntax for the INSD entries is discussed among the three databanks at an INSD collaborative meeting held once every year. The agreed rules are reflected in feature tables that define the common syntax (http://www.ddbj.nig.ac.jp/FT/full_index.html).
In the last year, we started novel web services that focus on daily announcements using Really Simple Syndication (RSS) technology and visualization of DDBJ content with high readability. Furthermore, a new data archive database for massive amounts of raw sequencing reads from next-generation sequencers was officially launched. The expert annotators of the DDBJ Read Archive (DRA) issue original accession numbers for submitted data. Concurrently, there was a preliminary release of a raw read annotation pipeline tool. This analytical pipeline tool supports reference genome mapping, de novo assembly and further annotation analyses, such as single nucleotide polymorphism (SNP) detection. The following sections describe three major advancements of the DDBJ databases, the novel announcement web services and the new archiving database with analytical tools for raw sequencing reads.
DEVELOPMENT OF DDBJ DATABASES
We have introduced newly released DDBJ databases, databases within the framework of INSD and other individual databases that have been appended from last year’s report (1).
Datasets contributing to INSD through DDBJ
In the period from July 2008 to June 2009, DDBJ collected and released original data on 1 701 110 entries/1 116 138 614 bases. More than 90% of the data came from Japanese researchers and the Japan Patent Office (JPO) and the rest was mainly from researchers in China, Korea and Taiwan. We call this dataset the ‘INSD-core data’. It consists of INSD data in traditional format and includes general sequence data, complete genomes, expressed sequence tags (ESTs), etc., but excludes whole-genome shotgun (WGS), mass sequence for genome annotation (MGA) and third party annotation (TPA). Large sets of contigs (i.e. overlapping reads) and finished sequences without annotation from an ongoing genome project can be submitted to INSD as WGS data. DDBJ has released one WGS entry (2 878 428 bp) on Staphylococcus aureus ssp. aureus Mu50-omega and 23 675 009 MGA entries (80 069 915 counts). All of these INSD-core, WGS and MGA data were collected, reviewed and accessioned by DDBJ. Another portion of the INSD-core data contains the complete genome sequence of an endosymbiont within protist cells in the gut of the termite (Candidatus Azobacteroides pseudotrichonymphae genomovar. CFP2) submitted by Institute of Physical and Chemical Research (RIKEN) and National Institute of Genetics; full-length cDNA (HTC) and EST entries for the tomato (Solanum lycopersicum) submitted by Kazusa DNA Research Institute; Genome Survey Sequences entries for the rat (Rattus norvegicus LE/Stm) submitted by Kyoto University; EST entries for rhizomes of Chinese liquorice (Glycyrrhiza uralensis) submitted by Chiba University, MGA entries for the human and mouse submitted by RIKEN Omics Science Center; and MGA entries for small RNAs of the silkworm (Bombyx mori) submitted by the University of Tokyo. These data can be obtained at the DDBJ ftp site (http://www.ddbj.nig.ac.jp/ftp_soap-e.html). The reader may find it worthwhile to refer to the two sets of data on the complete genome sequence of an endosymbiont within protist cells in the termite gut and the MGA datasets used in Functional annotation of the mammalian cDNA (FANTOM; http://fantom.gsc.riken.jp/). This bacterial endosymbiont is widely known as a model organism for the study of cellulolysis. With regard to the endosymbiont, functional annotation of the bacterial genome has revealed that nitrogen fixation and cellulolysis are coupled within the protist’s cells (2). An MGA dataset from the FANTOM consortium identified a large-scale gene network that controls the differentiation of the human myeloid leukaemia cell line THP-1 from monoblast to monocyte by applying next-generation sequencing technology and the Cap Analysis Gene Expression (CAGE) method (3).
Datasets released from DDBJ
In Table 1, we summarize numbers of published records collected and released from DDBJ. A primary database is a database as originally constructed and a secondary database is based on a primary database. An MGA is defined as a sequence that is produced in large quantity for the purpose of genome annotation, such as CAGE and 5′SAGE. A TPA (4) is a nucleotide sequence data collection in which each entry is obtained by assembling primary entries publicized from INSD and/or the Trace Archive with additional feature annotations determined by experimental or inferential methods by the TPA submitter. The DDBJ Trace Archive (DTA) is a permanent repository of DNA sequence chromatograms (traces), base calls and quality estimates for single-pass reads from various large-scale sequencing projects. The DTA has operated since 2008. In 2009, a simple metadata search system and a viewer of trace data for DDBJ-accepted data were added. Gene Trek in Prokaryote Space (GTPS) (5) is a database of prokaryotic genome data that have been reannotated by analyzing the original data in various ways. Genome Information Broker (GIB) (6) is a comprehensive data repository of complete microbial genomes in the public domain. GIB distributes genome sequence data and annotation 1 day after the data are submitted to INSD. The DDBJ Amino Acid Sequence Database (DAD) is produced by extracting all translated sequences from the DDBJ periodical release, including all INSD (DDBJ/EMBL-Bank/GenBank) entries. We also support two other databases by providing maintenance service: Center for Information Biology gene Expression database (CIBEX) (7) is a public database for microarray data and stores MIAME-compliant data in accordance with MGED Society recommendations; Genomes TO Protein structures and function (GTOP) (8) is a database consisting of data analyses of proteins identified by various genome projects. The GTOP database mainly uses sequence homology analyses and features extensive use of information on 3D structures.
Table 1.
Datasets released from DDBJ
| Type | Database name | No. of records | Released date |
|---|---|---|---|
| Primary DB | INSD-core (processed by DDBJ) | 17 440 910 entries (1 701 110 entries) | 29 May 2009 |
| WGS | 1 246 513 entries | 10 September 2009 | |
| MGA | 34 740 058 entries | 1 June 2009 | |
| TPA | 593 entries | 10 September 2009 | |
| DTA | 2 submissions | 7 July 2008 | |
| DRA | 12 submissions | 11 September 2009 | |
| Secondary DB | DAD | 14 710 673 entries | 29 May 2009 |
| GTPS | 690 genomes | 25 May 2009 | |
| GIB | 982 genomes | 10 September 2009 |
The number of records represents only published data.
DAILY RELEASE ANNOUNCEMENT AND COMPREHENSIVE VISUALIZATION OF DDBJ DATABASES
To deliver up-to-date information from DDBJ to researchers every day, we started the daily publication of newly released data from DDBJ by implementing the following two new functions into the DDBJ web services. The first function is the announcement of RSS feeds of contents of data released from DDBJ databases each day (Figure 1). The second function is the visualization of DDBJ entries as word cloud figures. The following sections explain these in detail.
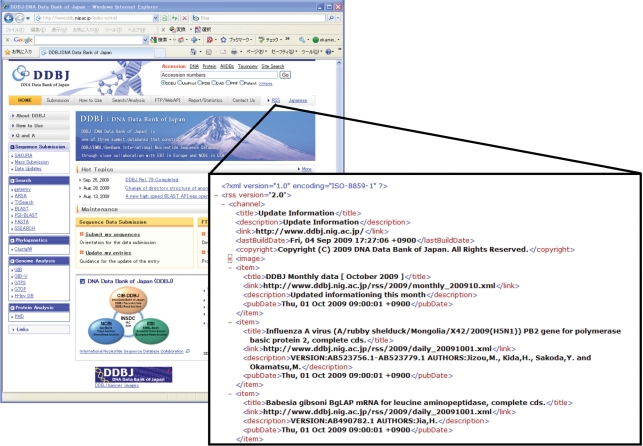
Figure 1.
A feed file for RSS 2.0 is published from the DDBJ homepage every day (http://www.ddbj.nig.ac.jp/rss/update_information.xml). Daily released contents of DDBJ databases can be confirmed via RSS reader programs.
Frequent announcement of daily data releases by RSS
The first new function is the RSS publication of daily data releases by DDBJ. The RSS is a family of web-feed formats used to publish frequently updated items such as blog entries and news headlines (9). RSS feeds are also used by biological databases such as PubMed Central (http://www.pubmedcentral.nih.gov/) and ArrayExpress (10). A list of new enhancements in FLATFILE/WGS/CON/TPA is generated as RSS feeds every day. The contents of the RSS feeds are generated in connection with the respective VERSION, ACCESSION ID, DEFINITION if these are present in REFERENCE tags. The unit of published content is set by PROJECT of DBLINK; however, if there are no XML tags in PROJECT, the TITLE of the REFERENCE tag and the AUTHOR are substituted for the PROJECT.
Comprehensive visualization of DDBJ entries by word cloud images
In addition to daily publication of database updates, information on classified statistics in DDBJ databases such as species and features is worthwhile for users. DDBJ already provides several statistics on its web site, such as the gross numbers of registered entries and of bases in registered databases, with numerical values and graphs. However, with conventional media it is difficult to provide an overview of the features of DDBJ databases at a glance. Therefore, we apply the word cloud image program Wordle (http://www.wordle.net/) to statistics on the frequency of DDBJ database. This program generates a word cloud image based on the frequency of keywords appearing in a text document or webpage. Figure 2 shows word cloud images in which the size of each word indicates its frequency, using keywords ranking among the top 100. To generate the figures, frequencies of species names and feature keys were calculated based on DDBJ release 78 of June 2009. Among the top 100 species, Homo sapiens occupies most of the image. On the other hand, the image for feature tags indicates extremely high frequencies of db_xref key and moderately high frequencies of product, protein_id, gene and translation. The keywords codon start, transl_table, note, mol_type and organism are also highlighted; the frequent words are feature keys for protein-coding sequences. These word cloud figures enable us to comprehensively capture information on the released DDBJ data at a glance.
Figure 2.
Word cloud images created using a DDBJ database release. The upper figure uses feature keys ranking among the top 100 for the total number of nucleotides; similarly, the lower figure uses species names.
DRA: A NEW DATABASE FOR NEXT-GENERATION SEQUENCERS
Overview of DRA
Next-generation sequencing platforms are revolutionizing biological science. These instruments are producing vastly more sequencing data than was ever possible with capillary technology. In addition, instead of microarrays, new sequencing platforms are used to measure molecular abundance because of their higher resolution and accuracy. In 2007, NCBI started the Short Read Archive (SRA) to accommodate the data from next-generation sequencing platforms. Early in 2008, EBI began operating the European Read Archive (ERA), and late in the same year DDBJ started to accept sequencing data from next-generation technologies such as Roche-454 Life Sciences GS FLX, Illumina Genome Analyzer and Applied Biosystems SOLiD. Initially, we prepared submission files at DDBJ and uploaded them to SRA. Since May 2009 we have operated a new repository, the DRA (http://trace.ddbj.nig.ac.jp/dra/index_e.shtml), to archive raw output data from new platforms. In June 2009, we started to issue our own internationally recognized accession numbers with prefix ‘DR’. Most submissions are from Japan. DRA has released 12 submissions by FTP and these data can also be retrieved from SRA. Considering the number of next-generation machines running in Japan and other Asian countries, the number of submissions to DRA is expected to increase.
Data model and validation system for DRA metadata
DRA uses the same metadata formats as SRA and ERA, and provides common accessions of the Submission (DRA), Study (DRP), Experiment (DRX), Sample (DRS) and Run (DRR) metadata objects with the prefix indicated in parentheses followed by a six-digit number (e.g. DRA000001). We are developing a submission system for DRA to improve submission throughput. As a first step, we have developed a web system, DRA Meta Checker, to validate metadata in XML file format (http://trace.ddbj.nig.ac.jp/DRAMetaChecker). This checker first validates uploaded XML files against an SRA XML schema, and then validates what cannot be validated by the schema, such as reference integrity among the XML documents, and correspondence between taxonomy ID and organism name. Detailed error, warning and usage messages are displayed after the validation process to help users create their metadata by themselves.
Data submission to DRA
We have released Excel spreadsheets for metadata submission to DRA, called ‘DRA sheets’ (Figure 3). Submitters are able to create metadata files by simply filling in the fields of familiar Excel files. Submitters can use the DRA sheets for three major platforms: 454, Genome Analyzer and SOLiD. Every field is explained by pop-up comments, required and optional fields are distinguished by colour, and the fixed fields contain entered values. In addition, these DRA sheets contain an Excel macro to generate the metadata XML files. Submitters can submit their metadata either in Excel file format or in XML file format (they can be validated by the DRA Meta Checker) as they prefer. For data transfer, submitters can use the FTP service of DDBJ or send a hard disk by a return-paid courier service. Once files have been received, the DRA team validates, issues accessions and uploads the data to SRA. DRA works with large sequencing centres producing massive amounts of data to establish a high-throughput submission pipeline between the centre and DRA.
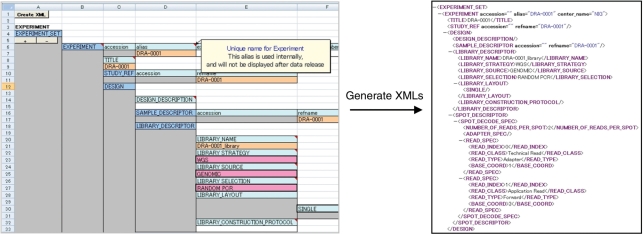
Figure 3.
DRA sheets: it contains an Excel macro to generate XML-formatted files for submission of metadata to DRA.
Planned development of DRA
At this moment, DRA is developing data release and retrieval systems, where they are currently supported as SRA systems. We will integrate the validation, submission creation and data transfer systems into a single fully automated interactive submission system to accommodate increased numbers of submissions. In May 2009, DDBJ/EBI/NCBI held its first international collaborative meeting on sequencing data from next-generation platforms. At this meeting, three databanks agreed to position the DRA/ERA/SRA activities within the framework of INSD and to prepare announcement articles for the research community and journal offices. DRA/ERA/SRA also discussed and agreed to develop a roadmap for XML schema releases with proposals for features, to establish a release policy, and to exchange (at least) metadata and FASTQ (sequence and quality values) data. DRA/ERA/SRA will collaborate to archive the data and share an accession space to provide a worldwide archive.
DDBJ READ ANNOTATION PIPELINE: AN ANALYTICAL TOOL FOR DRA
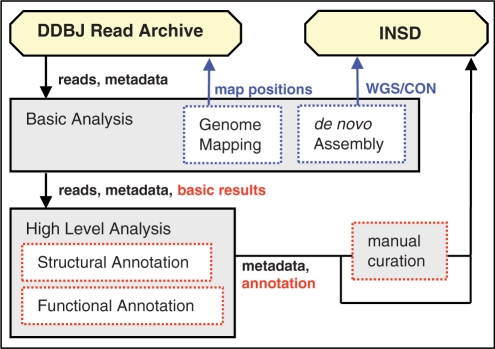
Automatic tools for the analysis of raw sequencing reads registered in DRA may be convenient and valuable for experimental biologists. We have developed a read annotation pipeline tool to annotate DRA-registered raw sequencing reads with high throughput. The ‘DDBJ Read Annotation Pipeline’ uses input data from FASTQ-formatted files in the DRA databases. The pipeline consists of two subprocesses: basic analysis for reference genome mapping and de novo assembly, and high-level analysis for combining automatic and manual annotations, such as SNP detection and expression tag counts (Figure 4).
Figure 4.
Flowchart of DDBJ Read Annotation Pipeline. The files of analytic results for structural and functional annotations are deposited in DDBJ databases, DRA and INSD.
The DDBJ Read Annotation Pipeline has the following three features. First, there is a short cut for the submission of analytical results to DDBJ databases, which means that map/assembly outputs are converted to DRA formats or DDBJ-based INSD formats. The second feature is high throughput, achieved by the use of a cluster computing system in DDBJ. The third feature is flexibility to select appropriate analytical tools from multiple candidates.
As a preliminary step for high-level annotation, analytical tools for SNP detection have been implemented in the current pipeline system. Other annotation tools, such as the high-level step, will be connected to the basic part. In general, to analyse massive amounts of raw reads requires high-level bioinformatics expertise. On the other hand, the DDBJ Read Annotation Pipeline enables experimental biologists to obtain results of automatic annotations by simply manipulating a graphical user interface. Currently, the pipeline only has the function of automatic annotation. To screen automatically annotated results, manual curation is indispensable [e.g. (11)]. Therefore, a user support function for further manual curation will be added to the pipeline tool.
FUTURE DIRECTIONS
In this report, we introduce the new archive database—the DRA—and an analytical pipeline for massive amounts of raw sequencing reads produced from next-generation sequencers. In the next step, we will integrate DRA, the pipeline and other automatic submission systems for DDBJ databases. The integrated framework will provide easier user access to the DDBJ databases.
FUNDING
DDBJ is funded by the Ministry of Education, Culture, Sports, Science and Technology of Japan with a management expenses grant for national university cooperation. The DRA and DTA are supported partially by the Integrated Database Project (http://lifesciencedb.jp/en) of the Database Center of Life Science in Japan. Funding for open access charge: The DDBJ management expenses grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We gratefully acknowledge the support of all members of DDBJ for data collection, annotation and release and for software development. In particular, we thank Takako Mochizuki and Dr Satoshi Saruhashi for constructing DRA and the pipeline, and Dr Satoshi Fukuchi, Dr Kazuho Ikeo, Prof. Hideaki Sugawara and Prof. Yoshio Tateno for support in the form of database maintenance and INSD collaboration.
REFERENCES
- 1.Sugawara H, Ikeo K, Fukuchi S, Gojobori T, Tateno Y. DDBJ dealing with mass data produced by the second generation sequencer. Nucleic Acids Res. 2009;37:D16–D18. doi: 10.1093/nar/gkn724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hongoh Y, Sharma VK, Prakash T, Noda S, Toh H, Taylor TD, Kudo T, Sakaki Y, Toyoda A, Hattori M, et al. Genome of an endosymbiont coupling n2 fixation to cellulolysis within protist cells in termite gut. Science. 2008;322:1108–1109. doi: 10.1126/science.1165578. [DOI] [PubMed] [Google Scholar]
- 3.FANTOM Consortium. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cochrane G, Bates K, Apweiler R, Tateno Y, Mashima J, Kosuge T, Mizrachi IK, Schafer S, Fetchko M. Evidence standards in experimental and inferential INSDC third party annotation data. OMICS. 2006;10:105–113. doi: 10.1089/omi.2006.10.105. [DOI] [PubMed] [Google Scholar]
- 5.Kosuge T, Abe T, Okido T, Tanaka N, Hirahata M, Maruyama Y, Mashima J, Tomiki A, Kurokawa M, Himeno R, et al. Exploration and grading of possible genes in 183 bacterial strains by a common fine protocol lead to new genes: gene trek in prokaryote space (GTPS) DNA Res. 2006;13:245–254. doi: 10.1093/dnares/dsl014. [DOI] [PubMed] [Google Scholar]
- 6.Fumoto M, Miyazaki S, Sugawara H. Genome information broker (GIB): data retrieval and comparative analysis system for completed microbial genomes and more. Nucleic Acids Res. 2002;30:66–68. doi: 10.1093/nar/30.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeo K, Ishi-i J, Tamura T, Gojobori T, Tateno Y. CIBEX: center for information biology gene expression database. C. R. Biol. 2003;326:1079–1082. doi: 10.1016/j.crvi.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 8.Kawabata T, Fukuchi S, Homma K, Ota M, Araki J, Ito T, Ichiyoshi N, Nishikawa K. GTOP: a database of protein structures predicted from genome sequences. Nucleic Acids Res. 2002;30:294–298. doi: 10.1093/nar/30.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winer D. RSS 2.0 Specification. 2003. http://cyber.law.harvard.edu/rss/rss.html.
- 10.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update–from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;30:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]