Abstract
Protein Analysis THrough Evolutionary Relationships (PANTHER) is a comprehensive software system for inferring the functions of genes based on their evolutionary relationships. Phylogenetic trees of gene families form the basis for PANTHER and these trees are annotated with ontology terms describing the evolution of gene function from ancestral to modern day genes. One of the main applications of PANTHER is in accurate prediction of the functions of uncharacterized genes, based on their evolutionary relationships to genes with functions known from experiment. The PANTHER website, freely available at http://www.pantherdb.org, also includes software tools for analyzing genomic data relative to known and inferred gene functions. Since 2007, there have been several new developments to PANTHER: (i) improved phylogenetic trees, explicitly representing speciation and gene duplication events, (ii) identification of gene orthologs, including least diverged orthologs (best one-to-one pairs), (iii) coverage of more genomes (48 genomes, up to 87% of genes in each genome; see http://www.pantherdb.org/panther/summaryStats.jsp), (iv) improved support for alternative database identifiers for genes, proteins and microarray probes and (v) adoption of the SBGN standard for display of biological pathways. In addition, PANTHER trees are being annotated with gene function as part of the Gene Ontology Reference Genome project, resulting in an increasing number of curated functional annotations.
INTRODUCTION
PANTHER (Protein ANalysis THrough Evolutionary Relationships) is a database of phylogenetic trees of protein-coding gene families from all kingdoms of life (1). Ancestral genes (representing most recent common ancestors of extant genes) are annotated with ontology terms describing gene function, and likely functional divergence events are identified and used to divide protein families into subfamilies of genes with similar function. Hidden Markov models (HMMs) are constructed for all families and subfamilies, which can be used for genome annotation projects, alone or as part of the InterPro database (2) that includes PANTHER as well as several other well-known protein annotation resources.
The main goal of PANTHER is to infer the evolution of gene function across as many genes in as many genomes as possible, and apply these inferences to predict the functions of genes that have not been directly characterized by experiment. In particular, there are large communities of researchers elucidating gene function for so-called ‘model organisms’ (e.g. those listed in Table 1) and these results provide a basis for inferring the functions of related genes in humans and other organisms. PANTHER applies both software tools and manual curation to perform these inferences as accurately as possible, and to keep them up-to-date as new experimental results accumulate. Gene function—or, more commonly, the function of gene products such as proteins—is described using terms from the Gene ontology (GO) (3,4), or from representations of molecular pathways.
Table 1.
Sources for complete sets of protein-coding genes in PANTHER version 7
| Organism or clade(s) | Five-letter code | Data source | Reference |
|---|---|---|---|
| Arabidopsis thaliana | ARATH | TAIR | (11) |
| Dicot plant | |||
| Caenorhabditis elegans | CAEEL | WormBase | (12) |
| Nematode worm | |||
| Danio rerio | DANRE | Ensembl, ZFIN | (13) |
| Zebrafish | |||
| Dictyostelium discoideum | DICDI | DictyBase | (14) |
| Cellular slime mold | |||
| Drosophila melanogaster | DROME | FlyBase | (15) |
| Fruit fly | |||
| Escherichia coli | ECOLI | EcoCyc | (16) |
| Bacterium | |||
| Gallus gallus | CHICK | Entrez Gene | (17) |
| Chicken | |||
| Homo sapiens | HUMAN | SwissProt | (18) |
| Human | |||
| Mus musculus | MOUSE | MGI | (19) |
| Mouse | |||
| Rattus norvegicus | RAT | RGD | (20) |
| Rat | |||
| Saccharomyces cerevisiae | YEAST | SGD | (21) |
| Budding yeast | |||
| Schizosaccharomyces pombe | SCHPO | GeneDB | (22) |
| Fission yeast | |||
| Other chordate genomes | Ensembl | (23) | |
| Other non-chordate genomes | Entrez Gene | (17) |
We have made several major modifications to the most recent version of PANTHER. One of the main developments is collaboration with the GO Consortium, in which PANTHER trees are being annotated with GO terms as part of the GO Reference Genome project (5). For PANTHER version 7, all previous associations of PANTHER subfamilies with function terms have been updated to GO terms. Ongoing annotation within the Reference Genome Project includes a complete evidence trail for inferred annotations all the way to the experimental results (literature articles) and evolutionary events upon which the inferences are based. Other important developments include improvements to the phylogenetic trees, inference of inter-species orthologs, inclusion of more genomes and support for several alternate database identifier types.
Improved hidden Markov Models and phylogenetic trees, and ortholog identification
Gene families covering fully sequenced genomes
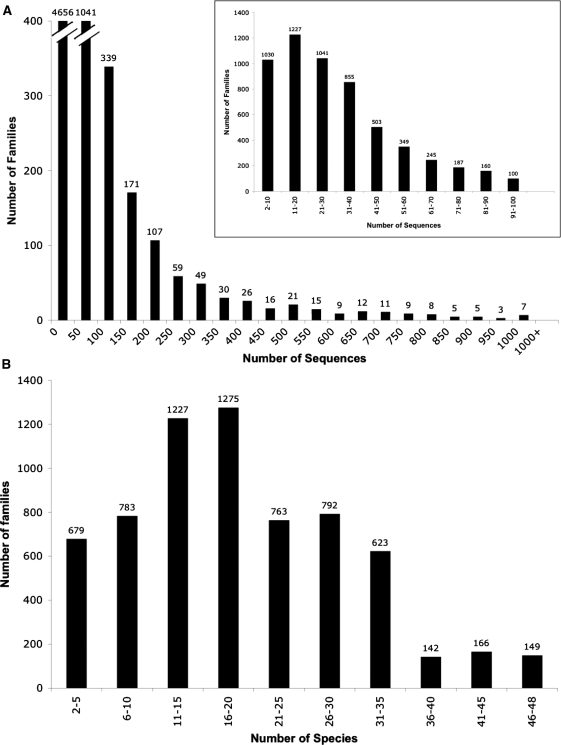
Previous versions of PANTHER focused on identifying subfamilies and the underlying functional divergence events. PANTHER 7 expands upon this focus by supporting accurate ortholog identification, and annotation of gene families ‘at any point in gene family evolution’, not just the major divergences. In order to meet these requirements, we made several important improvements to PANTHER. First, PANTHER trees aim to represent ‘all’ protein-coding genes from a phylogenetically diverse set of organisms. For PANTHER 7 trees, complete protein-coding gene sets for 48 different organisms were carefully constructed from a number of different sources, in collaboration with the GO Consortium, with an effort to use curated sources for model organism genomes (Table 1). These sets can be downloaded at ftp://ftp.pantherdb.org/genome/pthr7.0. We were careful to maintain stable PANTHER family and subfamily accession numbers from the previous version 6.1 to 7.0. To define protein family membership, each PANTHER 7 protein sequence was scored against the HMMs from version 6.1 and assigned to the family with the highest HMM score. If the resulting protein family contained over 1000 sequences, we attempted to manually divide it into smaller families to facilitate web browsing. We divided a total of 20 families from PANTHER 6.1, which have dramatically expanded due to numerous gene (or domain) duplication events, such as G protein-coupled receptors (GPCRs), ATP binding cassette (ABC) transporters, protein kinases, cytochrome P450s (CYP), and proteins containing ankyrin repeats, leucine-rich repeats (LRR), zinc finger and homeobox domains. Figure 1 shows the distribution of family sizes in terms of the number of distinct genes (Figure 1A) and the number of distinct genomes (Figure 1B) they contain.
Figure 1.
Distribution of protein family sizes in PANTHER version 7. (A) The distribution of the total number of genes (in all 48 genomes) per family. The N50 is about 150, i.e. about half the genes are in families larger than 150 members, and half are in smaller families. (B) The distribution of the total number of genomes per family. Most families contain genes from over 15 different species.
Improved multiple sequence alignments and HMMs
A multiple sequence alignment was constructed for each family using the MAFFT program (6) and a phylogenetic tree was estimated from the protein multiple alignment. Subfamily identifiers from version 6.1 were then ‘forward tracked’ to ancestral nodes in the version 7.0 trees whenever possible. In addition, in many cases, due to improvements in the phylogenetic trees in PANTHER 7 (see below), subfamily boundaries were refined during manual curation. After manual review and correction, if necessary, of the locations of both forward tracked and new subfamilies, a new HMM was constructed for each family and subfamily. We modified our existing HMM construction process (7) to make use of the multiple alignment from MAFFT. For PANTHER 7, we took the relevant sequences in the MAFFT alignment, trimmed it to include as match states only those columns aligned by ≥30% of the sequences in the subalignment [sequences were weighted using the same technique as in (1)], and used it to construct an initial model using the modelfromalign program in SAM3.1. We then used this initial model as input, in addition to the sequences themselves, to the buildmodel program using the same parameters as in (7). As a result, unlike in previous versions of PANTHER, the HMMs can have different lengths for different subfamilies, and now model any domains that are conserved across a single subfamily but not found in other subfamilies.
New algorithm for phylogenetic trees
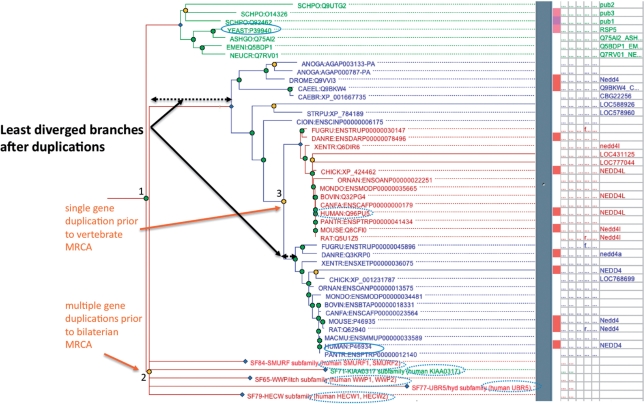
PANTHER trees aim to accurately represent ‘all’ of the evolutionary events in the gene family; for PANTHER 7, this means accurately inferring speciation and gene duplication events. For the gene trees, we use a novel algorithm, GIGA (Gene tree Inference in the Genomic Age). GIGA makes use of the known species tree and the presumably complete gene sets to infer accurate gene trees and locate gene duplication events relative to speciation events. If more than one gene duplication event took place between given consecutive speciation events, this appears as a single, multifurcating duplication node (e.g. node ‘2’ in Figure 2). The algorithm also performs a fast, approximate reconstruction of ancestral protein sequences at each node in the tree, using an iterative procedure starting at the leaves of the tree (modern day sequences) that considers the descendant sequences and the nearest outgroup.
Figure 2.
Example of human orthologs and LDO of the yeast RSP5 gene, identified using a phylogenetic tree. The figure shows part of the tree for PTHR11254 (HECT domain ubiquitin–protein ligase family), tracing the evolutionary relationship between RSP5 and its orthologs in humans, particularly its LDO, NEDD4. Orange nodes represent gene duplication events, green nodes represent speciation events, blue nodes represent subfamily nodes; in this figure blue nodes represent genes present in the bilaterian common ancestor that went on to found subfamilies. The solid outline ovals indicate the LDO pair in human and yeast, RSP5 and NEDD4 respectively. RSP5 has an additional nine orthologs in humans (dashed-outline ovals), but these have diverged to a greater degree than NEDD4. Conversely, 10 human genes have RSP5 as the ortholog, but only NEDD4 has RSP5 as the LDO. The LDO is identified by starting with the MRCA, and following the branch with the shortest length (least sequence divergence) after each gene duplication event. In this example, the MRCA is the speciation event that separated NEDD4 from RSP5 (labeled ‘1’), and there are at least two gene duplication events in the NEDD4 lineage: one at the base of the bilaterians representing multiple events that occurred in relatively rapid succession (labeled ‘2’) to create six genes in total and one at the base of the vertebrates (labeled ‘3’) to create the ancestors of NEDD4 and NEDD4L.
Orthologs: identification of complete set of orthologs and best one-to-one (least diverged) ortholog
These improved gene trees provide the basis for accurate inference of orthologs, pairs of genes whose most recent common ancestor (MRCA) diverged due to a speciation event (8). Orthologs of each gene can be viewed on PANTHER gene pages, and the entire set of pairwise ortholog inferences can be downloaded from the PANTHER website (http://www.pantherdb.org/downloads). For orthologs, PANTHER reports not only one-to-one but also one-to-many (i.e. when gene duplication has occurred in one lineage following speciation) and many-to-many orthologs (i.e. when gene duplication has occurred in both lineages following speciation). In the case of multiple orthologs, PANTHER identifies the one-to-one relationship that has ‘diverged the least’ following any gene duplication events. The ‘least diverged ortholog’ (LDO) pairs therefore represent the most nearly ‘equivalent’ gene pairs between different organisms based on the phylogenetic tree. Following gene duplication, the most common fates of the copies are thought to be neofunctionalization (in which one copy retains the ancestral function, while the other adapts to a new function) and subfunctionalization (in which each copy specializes in a subset of the ancestral functions) (9). If neofunctionalization has occurred, the LDO is the copy predicted to retain the ancestral function, i.e. the ‘same gene’ as the ancestor. An example of ortholog and LDO identification is shown in Figure 2.
Expanded sets of genomes and sequence identifiers for PANTHER tools
Since its inception, the PANTHER website has provided, for a limited set of ‘fully supported’ genomes (human, mouse, rat and fruit fly), the following functionality: (i) stored classifications for all protein-coding genes, including family, subfamily, molecular function, biological process and pathway, (ii) visualization tools such as the whole genome pie chart view (Figure 3) of gene functions and (iii) analysis tools such as the Gene Expression Analysis Tool (10) for analyzing user-generated data relative to PANTHER classifications. For version 7, we have increased the number of fully supported genomes from 4 to 12 organisms, those participating in the GO Reference Genome Project (5), listed at the beginning of Table 1.
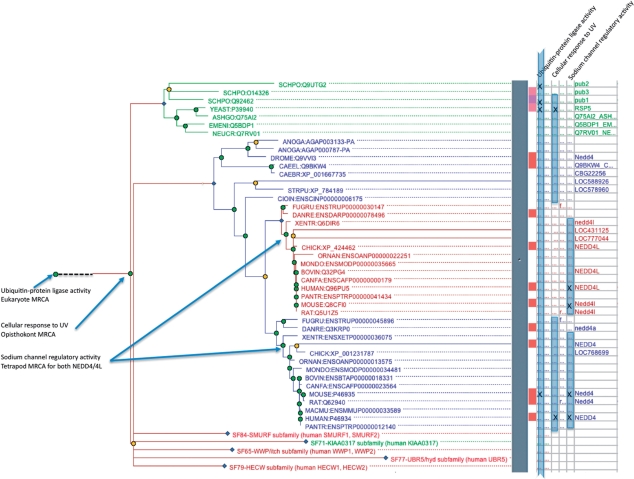
Figure 3.
Annotating a PANTHER tree with GO terms, and inferring GO terms for other genes by homology. The tree is the same as in Figure 2. The ‘x’ marks in the adjoining table (right panel) show the experimental GO annotations for each gene in the tree. For instance, yeast RSP5 has been determined experimentally to have the function ‘ubiquitin–protein ligase activity’, and be involved in the process of ‘cellular response to UV’. Based on the distribution of experimental annotations among genes, and, in some cases, the target of protein activity, one can infer annotations of ancestral genes. For instance, yeast RSP5 and human NEDD4 have been experimentally determined to operate in ‘cellular response to UV’, through targeting of the RNAPII protein for degradation, so this function was likely present in their common ancestor and inherited by descent from this ancestor. PANTHER captures this ancestral gene annotation, as well as rules for inferring functions for experimentally unannotated genes (shown with blue bars). In this example, the ancestral gene annotation allows us to infer ‘cellular response to UV’ for all least-diverged orthologs of NEDD4/RSP5 in animals and fungi. Note that different function annotations are inferred to have arisen in different ancestral genes (annotated nodes at left); this results in different inferred annotations across the genes in the family (blue bars indicating gene annotations at right). For instance, all genes in the tree can be inferred to have ‘ubiquitin–protein ligase activity’, while only a few genes (tetrapod orthologs of human NEDD4 and NEDD4L) can be inferred to have ‘sodium channel regulatory activity’ (as their targets, specific epithelial sodium channel subunits, apparently evolved first in tetrapods, not shown).
In addition, we have increased the number of different database identifiers supported by PANTHER tools and in searches of the PANTHER database. Previously, for genes only identifiers from NCBI Entrez Gene (17) or FlyBase (15) were supported; for proteins only RefSeq (24) or FlyBase identifiers. In PANTHER 7, we now also support identifiers from Ensembl (23), model organism databases, the International Protein Index (IPI) (25) and UniProt (18). All of these identifiers are obtained through the mapping files provided by UniProt (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/idmapping/).
Pathway diagrams using SBGN
PANTHER 7 has adopted the Systems Biology Graphical Notation (SBGN) standard (26) for the 165 pathway diagrams currently available on the PANTHER website. This standard was recently released at http://sbgn.org and provides a consistent semantics for symbols used in pathway diagrams.
Collaboration with GO Consortium
For almost 2 years now, there has been a formal collaboration between the Gene Ontology Consortium and the PANTHER database (5). As a result, in PANTHER 7, all molecular function, biological process and cellular component terms are exclusively GO terms [previous versions of PANTHER used the PANTHER/X ontology (1), though a mapping file to GO was provided]. The PANTHER/X biological process ontology has been retired, but we have retained the PANTHER/X molecular function ontology and renamed it ‘Protein Class’ since many terms are quite different from those in GO, and we have gotten considerable feedback from users about its utility.
As part of the GO Reference Genome Project, GO curators are annotating trees from the PANTHER database with GO terms describing molecular function, biological process and cellular component. As described in (5), the goal of this project is to provide accurate, complete and consistent GO annotations for all genes in 12 model organism genomes. GO terms based on experimental data from the scientific literature are used to annotate ancestral genes in the phylogenetic tree; thus, unannotated descendants of these ancestral genes are inferred to have inherited these same GO annotations by descent. An example of this annotation process is shown in Figure 3.
This rigorous process for evolutionary inference provides a means for accurate inference of GO annotations by homology, as well as a means for comparing and consistency-checking annotations for related genes. While earlier versions of PANTHER have allowed annotation of ‘subfamily nodes’ (i.e. ancestral genes that founded a particular subfamily), this more generalized GO annotation process requires all ancestral genes to be annotatable in principle, which has only become supported with the release of PANTHER 7. For most end users, perhaps the most relevant outcomes of this collaboration will be: (i) an increased number of GO annotations, especially those inferred by homology and (ii) the ability to trace all of the evidence behind each homology-based annotation. This evidence includes not only the gene that was experimentally demonstrated to perform a particular function (and the scientific publication reporting the experiment), but also the ancestral gene in which the function was inferred to have evolved. In the long term, all PANTHER ontology annotations will be migrated to this new standard.
FUNDING
National Institute of General Medical Sciences (GM081084). Funding for open access: SRI International.
Conflict of interest statement. None declared.
REFERENCES
- 1.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaudet P, Chisholm R, Berardini T, Dimmer E, Engel S, Fey P, Hill D, Howe D, Hu J, Huntley R, et al. The Gene Ontology's; Reference Genome Project: a unified framework for functional annotation across species. PLoS Comput. Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinformatics. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 7.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 9.Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20:544–549. doi: 10.1016/j.tig.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogers A, Antoshechkin I, Bieri T, Blasiar D, Bastiani C, Canaran P, Chan J, Chen WJ, Davis P, Fernandes J, et al. WormBase 2007. Nucleic Acids Res. 2008;36:D612–D617. doi: 10.1093/nar/gkm975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe DG, Knight J, et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36:D768–D772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fey P, Gaudet P, Curk T, Zupan B, Just EM, Basu S, Merchant SN, Bushmanova YA, Shaulsky G, Kibbe WA, et al. dictyBase–a Dictyostelium bioinformatics resource update. Nucleic Acids Res. 2009;37:D515–D519. doi: 10.1093/nar/gkn844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bairoch A, Bougueleret L, Altairac S, Amendolia V, Auchincloss A, Argoud-Puy G, Axelsen K, Baratin D, Blatter MC, Boeckmann B. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE. The Mouse Genome Database genotypes::phenotypes. Nucleic Acids Res. 2009;37:D712–D719. doi: 10.1093/nar/gkn886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dwinell MR, Worthey EA, Shimoyama M, Bakir-Gungor B, DePons J, Laulederkind S, Lowry T, Nigram R, Petri V, Smith J, et al. The Rat Genome Database 2009: variation, ontologies and pathways. Nucleic Acids Res. 2009;37:D744–D749. doi: 10.1093/nar/gkn842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 26.Le Novere N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM, et al. The Systems Biology Graphical Notation. Nat. Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]