Abstract
The RegPrecise database (http://regprecise.lbl.gov) was developed for capturing, visualization and analysis of predicted transcription factor regulons in prokaryotes that were reconstructed and manually curated by utilizing the comparative genomic approach. A significant number of high-quality inferences of transcriptional regulatory interactions have been already accumulated for diverse taxonomic groups of bacteria. The reconstructed regulons include transcription factors, their cognate DNA motifs and regulated genes/operons linked to the candidate transcription factor binding sites. The RegPrecise allows for browsing the regulon collections for: (i) conservation of DNA binding sites and regulated genes for a particular regulon across diverse taxonomic lineages; (ii) sets of regulons for a family of transcription factors; (iii) repertoire of regulons in a particular taxonomic group of species; (iv) regulons associated with a metabolic pathway or a biological process in various genomes. The initial release of the database includes ∼11 500 candidate binding sites for ∼400 orthologous groups of transcription factors from over 350 prokaryotic genomes. Majority of these data are represented by genome-wide regulon reconstructions in Shewanella and Streptococcus genera and a large-scale prediction of regulons for the LacI family of transcription factors. Another section in the database represents the results of accurate regulon propagation to the closely related genomes.
INTRODUCTION
Genome-scale annotation of regulatory features and reconstruction of transcriptional regulatory networks (TRNs) in a variety of diverse microbes constitute an important (albeit essentially unmet) challenge of modern genomics and systems biology. Such annotation is a prerequisite for understanding molecular mechanisms of transcriptional regulation in prokaryotes, comparison of gene content and topology of TRNs in related species and construction of realistic models of TRN evolution (1,2). The major components of bacterial TRNs are transcription factors (TF), their target genes and TF-binding sites (TFBS) in upstream regulatory regions of the respective operons. Many TFs act on multiple genes that collectively constitute a regulon. All regulons taken together form a TRN of the cell. Genes and operons co-regulated by the same TF and sharing TFBS are considered to be a part of a regulon. TFs from more than 50 distinct protein families comprise around 5–10% of all genes in an average prokaryotic genome, and their respective regulons cover a substantial fraction of bacterial TRNs (3).
The existing web-resources, such as RegulonDB, DBTBS, CoryneRegNet, MTBregList and PRODORIC, collect experimental knowledge on transcriptional regulation mostly in model bacteria, such as Escherichia coli, Bacillus subtilis, Corynebacterium glutamicum and Mycobacterium tuberculosis (4–8). Another database, RegTransBase, developed by our group, contains published data on transcriptional regulation in a broader range of prokaryotic genomes (9). Several microbial databases based on in silico regulon reconstructions also exist. Tractor_DB provides an access to automatic genomic propagations of previously described regulons in E. coli regulons to a set of 30 genomes of other γ-proteobacteria (10). MycoRegNet represents the result of bioinformatic transfer of the well-examined TRN of C. glutamicum described in the CoryneRegNet database to M. tuberculosis (11).
A growing number of complete prokaryotic genomes promoted active development of comparative genomic approaches for prediction of cis-acting regulatory elements and regulon reconstructions. Major directions of this analysis involve (i) annotation and propagation of previously known TF regulons from model organisms to many others; and (ii) ab initio discovery and reconstruction of novel TF regulons providing novel regulatory annotations for a large number of genes in bacterial genomes [see a recent review (3) for a detailed description of strategies for comparative reconstruction of regulons]. During the past decade we have focused on in silico reconstruction and manual curation of various metabolic regulons across large sets of bacterial genomes using comparative genomic techniques (see ‘Database content’ section). In addition to reconstruction of regulons controlling a particular biological process, we initiated wide-ranging reconstructions of entire TRNs in several groups of closely related species [e.g. Shewanella (12)]. Based on this progress, we expect the number of other genome-wide regulon reconstructions for many other groups of prokaryotes to be growing at the accelerated pace.
To provide public access to the results of these studies, and to promote further analysis, validation and modeling of reconstructed TF regulons we designed the RegPrecise database. In contrast to other databases dealing with transcriptional regulation in bacteria, regulon descriptions in the RegPrecise database result from comparative genomic reconstruction and thorough manual curation in a significant number of species and thus constitute large-scale and high-quality regulatory annotations.
DATABASE CONTENT
The RegPrecise contains high confidence regulatory annotations (both published and unpublished) obtained by careful comparative genomic analysis and manual curation of each regulon included in the dataset. These manually curated regulon reconstructions constitute the major section in the database. The second part of the database represents accurate automatic propagation of manually predicted regulons to the large set of closely related genomes.
Manually curated regulons
These annotation are generated using the approach of Mironov and co-authors (13) based on simultaneous analysis of transcription factor binding sites in several related genomes. Main assumption of this approach is that true TF binding sites are at least partially conserved in evolution, whereas false positive sites are mostly not conserved and randomly scattered even in closely related genomes (14). Over the past decade, a similar approach was used for de novo identification and reconstruction of various metabolic regulons in a number of diverse taxonomic groups of bacteria [reviewed in (3), see also (15–21)]. Examples of reconstructed regulatory networks in bacteria include regulons that control metabolism of vitamins and cofactors (22–24), amino acids and fatty acids (25–27), utilization of carbohydrates (28,29), metal homeostasis (30–32) and response to anaerobiosis (33,34). Many components of regulatory subnetworks, such as the iron-responsive TRN in α-proteobacteria (31), the nitrogen oxide–responsive TRN in diverse bacteria (34), the sulfate reduction regulon HcpR in δ-proteobacteria (35) and the ribonucleotide reductase regulon NrdR in bacteria (36), were predicted by this approach and then confirmed experimentally by independent research groups [see references in (3)].
Recent availability of a large number of complete genomic sequences for several taxonomic groups of closely related bacteria provides opportunity to perform genome-wide reconstruction of their TRNs. Our pilot analysis of thirteen Shewanella genomes resulted in the reconstruction of TRN, which includes 74 TFs and 3110 TFBSs (12), revealing substantial differences compared to the classical E. coli model. In a recent analysis of eight representative Streptococcus genomes, we identified candidate TFBSs and reconstructed regulons for 30 known and predicted TFs. These and other previous studies helped us define a general workflow of the ‘knowledge-driven’ approach for genomic reconstruction of regulons and develop a concept of the RegPrecise database for collection and visualization of the accumulated regulatory reconstructions. The results of these studies are included in the first release of RegPrecise.
Automatically propagated regulons
This section of the database represents the results of accurate propagation of the manually curated regulons to novel closely related genomes. To propagate a particular regulog to a target genome, we first require the presence of ortholog of the regulog-related transcription factor in this genome. Once it is found, we perform search for candidate TF-binding sites in upstream regions of genes being orthologous to one of the previously described and manually curated members of the regulog. For this aim we used the regulog-specific site search profile with a minimal site score observed in the regulog chosen as a threshold. The results of application of propagation procedure are summarized in a table, where for each regulog and for each novel genome, the number of target operons with conserved TF-binding site is indicated. The suggested regulon propagation procedure is considered to be accurate and conservative, since it relies on the manually curated regulons and does not make an attempt for automatic prediction of new members of regulon. The propagation procedure was based on orthologs developed in MicrobesOnline database (39).
DATA ORGANIZATION
The RegPrecise database has a hierarchical structure of the data organized at three major levels: (i) regulon, (ii) regulog and (iii) collection of regulogs (Figure 1). A single regulon in a particular genome is a primary object of the database. A regulon has a clear biological interpretation as a set of genes in one organism that are co-regulated by a common TF. The regulon is characterized by a TF, its predicted DNA-binding site model (a profile), a set of target genes/operons together with associated TF-binding sites in their upstream regions. At the next level of hierarchy, RegPrecise uses a regulog concept (15) to represent a particular TF regulon inferred and projected in a set of closely related genomes. The regulog represents the main outcome of application of the comparative genomic analysis for TF regulon reconstruction in a group of genomes. TFs widely distributed across bacterial lineages can be linked to multiple lineage-specific regulogs with variable TFBS motifs. The regulog level allows the user to analyze conservation of regulon content across a group of genomes. The third and highest level the database is represented by collections of regulogs of three major types grouped by: (i) taxonomy, (ii) individual TF, and (iii) TF family and (iv) pathway or subsystem (according to the functional classification of regulated genes). Each of these types of regulog collections are briefly described below and illustrated by examples from the RegPrecise.
Figure 1.
Hierarchical data organization in the RegPrecise database. Major objects in the database are TFs, TFBSs and target genes. TFs are attributed to a certain protein family and effector. Target genes participate in a particular pathway or process. Three major levels in the database are: (i) regulon constituted by a set of genes co-regulated by TF in a single genome, (ii) regulog formed by a set of orthologous regulons in a group of related genomes and (iii) collection of regulogs by one of four categories (see text). Each TF regulog is linked to a unique TFBS motif.
Collection by taxonomic groups
Collection by taxonomic groups organizes all reconstructed TF regulogs for a given set of closely related genomes. Some of these collections represent results of large-scale reconstructions of regulons in narrow taxonomic groups of bacteria. These regulon collections are valuable for modeling transcriptional and metabolic networks and could be used as a framework for interpretation of high-throughput gene expression data in some model microorganisms. Currently the database contains two collections of this type obtained for the groups of 13 Shewanella spp. and 8 Streptococcus spp. that include 74 and 38 TF regulogs, respectively. We anticipate constant growth in the number of collections of this type, as more and more other well populated groups of closely related bacterial genomes become available.
Collection by Transcription Factors
Collection by transcription factors includes reconstructions of the orthologous TF regulons across different taxonomic groups. Currently the database features nine collections of regulons of this type including the Irr, IscR, LiuR, NiaR, NrtR, NrdR, PsrA, RutR and Zur regulons reconstructed in diverse microbial lineages. Each of these collections is composed by several lineage-specific regulogs (from 5 to 18 regulogs) that may have variable regulon content and somewhat diverged TFBS motifs. For instance, 18 lineage-specific NrtR regulogs have highly variable regulon content and diverged TFBS motifs that have a common GT-(N7)-AC consensus conserved only for 12 regulogs (22). In another example, 11 NrdR regulogs have mostly conserved sets of target genes (nrdAB, nrdJ, nrdDG) and weakly diverged TFBS motifs with a common consensus CAN-(N4)-TNG for all lineage-specific regulogs (36). The TF-based collections of regulogs provide a useful view for assessment of overall regulon conservation across taxonomic groups of bacteria enabling comparison and evolutionary analysis of their TFBS motifs.
Collection by TF family
Collection by TF family is similar to the collection by TFs but it includes large-scale results of regulog reconstruction for different regulators from the same TF protein family. Currently the database includes a single collection of ∼220 regulogs representing the LacI family of TFs. This collection covers ∼270 bacterial genomes and contains over 4800 TFBSs. The majority of TFs from the LacI family control various sugar utilization pathways. The front page for this collection provides a bird-eye view of the variability of TFBS motifs identified for the LacI-type TF regulogs that have only two generally conserved positions, G and C, in the middle part of most TFBS motifs. Such TF family-broad collections may provide a basis for systematic analysis of TFBS motifs evolution and covariation of nucleotides in these DNA motifs and amino acids in DNA-binding domains of TFs (37).
Collection by pathway or subsystem
Collection by pathway or subsystem combines all TF regulogs that control genes involved in the same metabolic or cellular process (pathway, subsystem). Currently this classification includes 10 categories: the metabolism of amino acids, cofactors, fatty acids, nucleotides, carbohydrates, nucleotides, nitrogen oxides, metal homeostasis, drug resistance, and stress response, and this functional coverage will be expanded in the future database updates.
A particular TF regulog can be simultaneously included in collections of several different types. For example, the LiuR–Shewanella regulog is present within three collections for: (i) the Shewanella taxonomic group; (ii) the LiuR TF; and (iii) the amino acid metabolism. We expect that any update of the database by novel TF regulogs will result in concurrent update of various types of TF regulog collections.
DATABASE ACCESS AND INTERFACE
The RegPrecise database is publicly accessible through a web interface at http://regprecise.lbl.gov. The home page provides several different ways to access the regulon descriptions. Two key entry points, ‘Regulon collections’ and ‘Browse and statistics’, allow browsing through the database content, whereas ‘Search gene/regulator’ is useful for finding information about specific target genes and TF regulators in individual genomes. Alternatively, in order to get an overview of the database content, two types of browsing are provided under ‘Browse and statistics’ link—‘Browse by regulog’ and ‘Browse by genome’.
Following ‘Regulon collections’ link, the user gets a list of all available collections of regulogs organized into groups corresponding to the four types of collections described above. Each collection web page provides condensed information about all TF regulogs inferred by the comparative genomics approach for a particular group of genomes, TFs, or biological pathways, and includes total statistics on a number of genomes, regulogs, TFs and TFBSs within a collection. Each type of collection is focused on certain aspects of evolution of transcriptional regulation, and thus requires a different way of the data representation. An interface implemented in the RegPrecise is illustrated below on two examples.
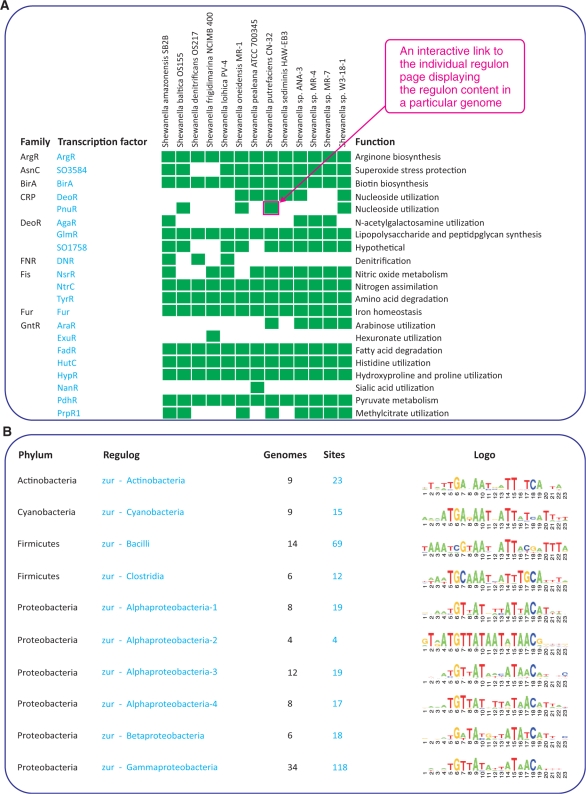
Representation of the collection of regulogs by taxonomic group (as illustrated by the ‘Shewanella’ collection in Figure 2A) provides an overview table of 74 reconstructed TF regulogs sorted by a TF protein family attribute in a set of 13 Shewanella genomes sorted by taxonomy. In this table, rows and columns correspond to regulogs and genomes, respectively, whereas each non-empty cell colored green provides a reference to a web page with detailed description of a particular TF regulon in individual genome. The table shows distribution of orthologous TFs in a group of genomes, highlights universally conserved and narrowly distributed regulogs, and provides general functional classification of target genes within the regulogs.
Figure 2.
Summary pages for collections of regulogs for a particular taxonomic group (A), and for a particular transcription factor (B). Collection of multiple TF regulons in the Shewanella species is displayed as a matrix of TFs versus genomes, where each green cell provides a link to species-specific regulon description (A). Collection of Zur regulogs in diverse taxonomic groups is represented as a table with respective TFBS motif logos.
Representation of the collection of regulogs by TFs (as illustrated by the Zur regulon collection in Figure 2B) provides a summary for all regulogs reconstructed for orthologous TFs across diverse taxonomic groups of bacteria. Each regulog has an attributed phylum name and the regulog name showing a more precise definition of the taxonomic group where it has been reconstructed. For this type of collections we also provide an alignment of TFBS motifs built using a set TFBSs inferred for each regulog. These TFBS motifs are represented by motif sequence logos drawn with the WebLogo package v.2.6 (38). Sequence logo is particularly useful for the comparison and evolutionary analysis of TFBS motifs between orthologous TF regulogs from different taxonomic groups.
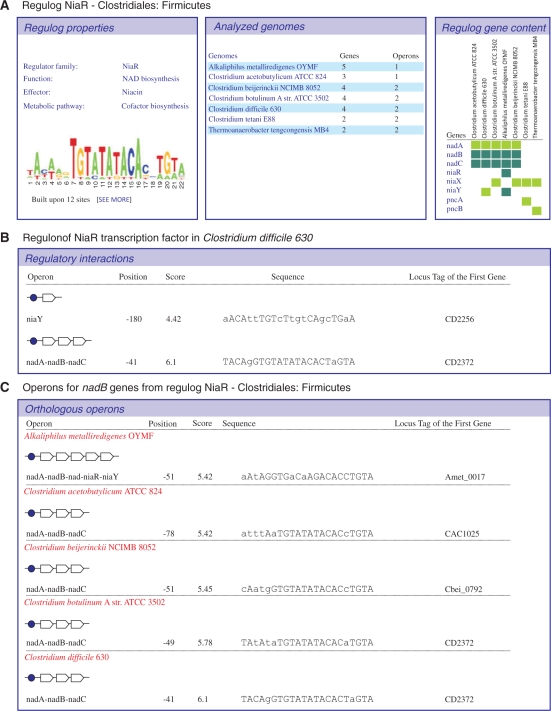
Regulog collection web pages, being the upper level in the data hierarchy of the RegPrecise, provide all necessary links to the web pages at the regulog and regulon levels. The regulog page provides a comparative table showing conservation of gene regulation across genomes within a particular regulog (Figure 3A). Essentially, this table shows a phylogenetic profile of gene regulation based on the presence and absence of gene regulation by a particular TF in every genome. This type of visualization allows the user to easily identify a core part of the regulon—a set of genes controlled by a TF in most of the analyzed genomes; and a variable part of the regulon populated by genes that are conserved only in several genomes. The regulog web page also provides a brief description of a TF (TF family, effector), a list of analyzed genomes with the number of predicted target genes and operons, and a TFBS motif sequence logo.
Figure 3.
Web representation of individual regulogs (A), regulons (B), and operons (C). Regulog page shows a summary, a TFBS motif sequence logo, a table with a set of analyzed genomes (with links to the genome-specific regulon pages), and a table of distribution of TF-regulated genes across these genomes (with links to the respective orthologous operon pages). Regulon page displays the list of regulated operons with detailed TFBS descriptions for a single genome (a horizontal view). Operon page provides a different view on orthologous regulated operons across multiple genomes.
The lower level in data hierarchy in the RegPrecise is a regulon described in the individual genome. The regulon page shows detailed information about all inferred regulatory interactions for a particular TF in a particular genome (Figure 3B). This web page has a brief description of a TF (Genbank locus tag, TF family, effector) and a complete list of predicted target genes organized in putative transcriptional units with detailed information about associated TFBSs (site sequence, score and position relative to the first gene start). In addition to this plain view on all target operons within a particular genome, we provide an orthogonal view on a particular operon in all genomes analyzed for a particular regulog (Figure 3C). The latter view allows the user to assess conservation of regulation for a particular operon.
Collections of regulogs, individual regulogs and regulon pages in the database are linked to the associated TFBS profile web pages that provide a list of all TFBSs identified for a particular regulog in a subset of genomes (including first gene locus tag, site sequence and relative position), and a TFBS profile represented as a sequence logo.
SUPPORTING EVIDENCES
A large number of regulon inferences previously described in bioinformatics papers and collected in the RegPrecise have been validated in targeted experiments. To provide the most up-to-date list of the related publications we organize the ‘Supporting evidences’ section with three subsections. ‘Publications’ provides a list of selected publications, where the comparative genomics approach was used for regulon prediction. ‘Experimental validations’ summarizes all regulogs for which at least one predicted regulatory interaction was experimentally validated (e.g. regulation of a target gene by a TF; validation of a TF-binding site). For 8 validated regulog we provide an updated set of publication references (overall 25 papers) with a brief description of investigated organisms and types of supporting experiment. We are planning to regularly update the information provided in the ‘Supporting evidences’ section, both for the regulons that were already deposited in the database and for any new regulon content. Finally, the ‘Recommended regulons for experimental verifications’ represents a list of predicted regulons recommended for future experimental testing. The latter web page may be valuable for experimental biologists who are looking for novel regulatory systems not previously characterized in any bacterial species.
FUTURE DEVELOPMENTS
Comparative genomic analysis and curation of regulons in groups of closely related bacterial genomes include several stages, both automatic and manual. We are working on a semi-automatic tool to significantly increase the speed of whole-genome regulon inference, but still retain their general quality level. We expect that regulogs obtained with this tool will be a major source of data in the RegPrecise database. Further we plan to add additional types of representation of regulatory information, in particularly for regulatory networks. This will improve the representation of regulons that have an overlapping set of target genes in a single genome. Also, we will add information about presence or absence of orthologous genes for every regulog member in ‘Regulon gene content’ tables on the respective regulog pages. Finally, we are working on connection of data on regulon pages in RegPrecise (TFs, regulated genes) with the microbial genome analysis resources such as MicrobesOnline (39), and community annotation resources such as SEED (40).
CONCLUSIONS
The RegPrecise database is an extensive collection of manually curated regulons inferred by the comparative genomics approach. In contrast to other regulatory databases, RegPrecise is not focused on a single model organism or a narrow taxonomic group, but provides a basis for comparative genomic reconstructions of regulons in many taxonomic groups. The RegPrecise visualizes the regulatory information with several interfaces developed to show many unique features of the proposed TF regulog collections at several hierarchical levels. In a few years we expect a fast growth in the comparative genomics data for regulons in bacteria due to the current fast growth in the number of well populated taxonomic groups of closely related genomes. We developed the RegPrecise database to face this oncoming challenge and be prepared to capture the massive results of semi-automatic regulon reconstructions by means of comparative genomics. The database also will serve as a platform for future high-throughput regulon validation using expression profiles of regulatory knockout mutants and by other approaches.
FUNDING
This work was part of the Virtual Institute for Microbial Stress and Survival (http://VIMSS.lbl.gov) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomics Program: GTL through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy; National Science Foundation (award DBI-0850546 to D.A.R.), Howard Hughes Medical Institute (55005610 to M.S.G.); Russian Fund for Basic Research (08-04-01000 to D.A.R. and 09-04-92745 to M.S.G.); Russian Academy of Sciences (program ‘Molecular and Cellular Biology’ to D.A.R and M.S.G.), Russian Science Agency (contract 2.740.11.0101 to M.S.G.), and Russian President’s grant for young scientists (MK-422.2009.4 to D.A.R.). Funding for open access charge: US Department of Energy (DE-AC02-05CH11231).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Andrey A. Mironov and the members of the Shewanella Federation for support, to Andrei L. Osterman and MicrobesOnline team for useful discussions and encouragement, and to Tatiana Smirnova for the artistic RegPrecise Web site design.
REFERENCES
- 1.Balleza E, Lopez-Bojorquez LN, Martinez-Antonio A, Resendis-Antonio O, Lozada-Chavez I, Balderas-Martinez YI, Encarnacion S, Collado-Vides J. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol. Rev. 2009;33:133–151. doi: 10.1111/j.1574-6976.2008.00145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gelfand MS. Evolution of transcriptional regulatory networks in microbial genomes. Curr. Opin. Struct. Biol. 2006;16:420–429. doi: 10.1016/j.sbi.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Rodionov DA. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 2007;107:3467–3497. doi: 10.1021/cr068309+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumbach J. CoryneRegNet 4.0 – a reference database for corynebacterial gene regulatory networks. BMC Bioinformatics. 2007;8:429. doi: 10.1186/1471-2105-8-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grote A, Klein J, Retter I, Haddad I, Behling S, Bunk B, Biegler I, Yarmolinetz S, Jahn D, Munch R. PRODORIC (release 2009): a database and tool platform for the analysis of gene regulation in prokaryotes. Nucleic Acids Res. 2009;37:D61–D65. doi: 10.1093/nar/gkn837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacques PE, Gervais AL, Cantin M, Lucier JF, Dallaire G, Drouin G, Gaudreau L, Goulet J, Brzezinski R. MtbRegList, a database dedicated to the analysis of transcriptional regulation in Mycobacterium tuberculosis. Bioinformatics. 2005;21:2563–2565. doi: 10.1093/bioinformatics/bti321. [DOI] [PubMed] [Google Scholar]
- 8.Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36:D93–D96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazakov AE, Cipriano MJ, Novichkov PS, Minovitsky S, Vinogradov DV, Arkin A, Mironov AA, Gelfand MS, Dubchak I. RegTransBase – a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 2007;35:D407–D412. doi: 10.1093/nar/gkl865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez AD, Espinosa V, Vasconcelos AT, Perez-Rueda E, Collado-Vides J. TRACTOR_DB: a database of regulatory networks in gamma-proteobacterial genomes. Nucleic Acids Res. 2005;33:D98–D102. doi: 10.1093/nar/gki054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krawczyk J, Kohl TA, Goesmann A, Kalinowski J, Baumbach J. From Corynebacterium glutamicum to Mycobacterium tuberculosis – towards transfers of gene regulatory networks and integrated data analyses with MycoRegNet. Nucleic Acids Res. 2009;37:e97. doi: 10.1093/nar/gkp453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, et al. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 13.Mironov AA, Koonin EV, Roytberg MA, Gelfand MS. Computer analysis of transcription regulatory patterns in completely sequenced bacterial genomes. Nucleic Acids Res. 1999;27:2981–2989. doi: 10.1093/nar/27.14.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelfand MS. Recognition of regulatory sites by genomic comparison. Res. Microbiol. 1999;150:755–771. doi: 10.1016/s0923-2508(99)00117-5. [DOI] [PubMed] [Google Scholar]
- 15.Alkema WB, Lenhard B, Wasserman WW. Regulog analysis: detection of conserved regulatory networks across bacteria: application to Staphylococcus aureus. Genome Res. 2004;14:1362–1373. doi: 10.1101/gr.2242604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conlan S, Lawrence C, McCue LA. Rhodopseudomonas palustris regulons detected by cross-species analysis of alphaproteobacterial genomes. Appl. Environ. Microbiol. 2005;71:7442–7452. doi: 10.1128/AEM.71.11.7442-7452.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erill I, Jara M, Salvador N, Escribano M, Campoy S, Barbe J. Differences in LexA regulon structure among Proteobacteria through in vivo assisted comparative genomics. Nucleic Acids Res. 2004;32:6617–6626. doi: 10.1093/nar/gkh996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guia MH, Perez AG, Angarica VE, Vasconcelos AT, Collado-Vides J. Complementing computationally predicted regulatory sites in Tractor_DB using a pattern matching approach. In Silico Biol. 2005;5:209–219. [PubMed] [Google Scholar]
- 19.Mwangi MM, Siggia ED. Genome wide identification of regulatory motifs in Bacillus subtilis. BMC Bioinformatics. 2003;4:18. doi: 10.1186/1471-2105-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wels M, Francke C, Kerkhoven R, Kleerebezem M, Siezen RJ. Predicting cis-acting elements of Lactobacillus plantarum by comparative genomics with different taxonomic subgroups. Nucleic Acids Res. 2006;34:1947–1958. doi: 10.1093/nar/gkl138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu M, Su Z. Computational prediction of cAMP receptor protein (CRP) binding sites in cyanobacterial genomes. BMC Genomics. 2009;10:23. doi: 10.1186/1471-2164-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodionov DA, De Ingeniis J, Mancini C, Cimadamore F, Zhang H, Osterman AL, Raffaelli N. Transcriptional regulation of NAD metabolism in bacteria: NrtR family of Nudix-related regulators. Nucleic Acids Res. 2008;36:2047–2059. doi: 10.1093/nar/gkn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodionov DA, Li X, Rodionova IA, Yang C, Sorci L, Dervyn E, Martynowski D, Zhang H, Gelfand MS, Osterman AL. Transcriptional regulation of NAD metabolism in bacteria: genomic reconstruction of NiaR (YrxA) regulon. Nucleic Acids Res. 2008;36:2032–2046. doi: 10.1093/nar/gkn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodionov DA, Mironov AA, Gelfand MS. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kazakov AE, Rodionov DA, Alm E, Arkin AP, Dubchak I, Gelfand MS. Comparative genomics of regulation of fatty acid and branched-chain amino acid utilization in proteobacteria. J. Bacteriol. 2009;191:52–64. doi: 10.1128/JB.01175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makarova KS, Mironov AA, Gelfand MS. Genome Biol. 2001. Conservation of the binding site for the arginine repressor in all bacterial lineages. 2, RESEARCH0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 2004;32:3340–3353. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodionov DA, Gelfand MS, Hugouvieux-Cotte-Pattat N. Comparative genomics of the KdgR regulon in Erwinia chrysanthemi 3937 and other gamma-proteobacteria. Microbiology. 2004;150:3571–3590. doi: 10.1099/mic.0.27041-0. [DOI] [PubMed] [Google Scholar]
- 29.Yang C, Rodionov DA, Li X, Laikova ON, Gelfand MS, Zagnitko OP, Romine MF, Obraztsova AY, Nealson KH, Osterman AL. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J. Biol. Chem. 2006;281:29872–29885. doi: 10.1074/jbc.M605052200. [DOI] [PubMed] [Google Scholar]
- 30.Panina EM, Mironov AA, Gelfand MS. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl Acad. Sci. USA. 2003;100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-proteobacteria. PLoS Comput. Biol. 2006;2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Permina EA, Kazakov AE, Kalinina OV, Gelfand MS. Comparative genomics of regulation of heavy metal resistance in Eubacteria. BMC Microbiol. 2006;6:49. doi: 10.1186/1471-2180-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravcheev DA, Gerasimova AV, Mironov AA, Gelfand MS. Comparative genomic analysis of regulation of anaerobic respiration in ten genomes from three families of gamma-proteobacteria (Enterobacteriaceae, Pasteurellaceae, Vibrionaceae) BMC Genomics. 2007;8:54. doi: 10.1186/1471-2164-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 2005;1:e55. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodionov DA, Dubchak I, Arkin A, Alm E, Gelfand MS. Reconstruction of regulatory and metabolic pathways in metal-reducing delta-proteobacteria. Genome Biol. 2004;5:R90. doi: 10.1186/gb-2004-5-11-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodionov DA, Gelfand MS. Identification of a bacterial regulatory system for ribonucleotide reductases by phylogenetic profiling. Trends Genet. 2005;21:385–389. doi: 10.1016/j.tig.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 37.Huang N, De Ingeniis J, Galeazzi L, Mancini C, Korostelev YD, Rakhmaninova AB, Gelfand MS, Rodionov DA, Raffaelli N, Zhang H. Structure and function of an ADP-ribose-dependent transcriptional regulator of NAD metabolism. Structure. 2009;17:939–951. doi: 10.1016/j.str.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alm EJ, Huang KH, Price MN, Koche RP, Keller K, Dubchak IL, Arkin AP. The MicrobesOnline Web site for comparative genomics. Genome Res. 2005;15:1015–1022. doi: 10.1101/gr.3844805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]