Abstract
The Gene Ontology (GO) Consortium (http://www.geneontology.org) (GOC) continues to develop, maintain and use a set of structured, controlled vocabularies for the annotation of genes, gene products and sequences. The GO ontologies are expanding both in content and in structure. Several new relationship types have been introduced and used, along with existing relationships, to create links between and within the GO domains. These improve the representation of biology, facilitate querying, and allow GO developers to systematically check for and correct inconsistencies within the GO. Gene product annotation using GO continues to increase both in the number of total annotations and in species coverage. GO tools, such as OBO-Edit, an ontology-editing tool, and AmiGO, the GOC ontology browser, have seen major improvements in functionality, speed and ease of use.
INTRODUCTION
The Gene Ontology (GO; http://www.geneontology.org) project is a major collaborative bioinformatics initiative that aims to standardize the representation of gene and gene product attributes across species. The project provides a controlled vocabulary of terms for describing gene product characteristics, supports gene product annotation data from GO Consortium (GOC) members, and develops tools to access and process these data. Over the past ten years, the GOC has expanded from its founding three model organism databases (mouse, yeast and fly) to include the world's; major repositories for plant, animal and microbial genomes. GOC makes its ontologies, annotations, and tools freely available to advance biological research.
ONTOLOGY DEVELOPMENT
The GO continues to mature in representing three aspects of biology, molecular functions (MF), biological processes (BP) and cellular components (CC). Table 1 illustrates the current contents of the GO website and database.
Table 1.
Current status of Gene Ontology as of 4 September 2009
| Biological process terms | 17 069 |
| Molecular function terms | 8637 |
| Cellular component terms | 2432 |
| Sequence ontology terms | 1603 |
| Annotation datasetsa | 52 |
| Species with annotation | 197 439 |
| Annotated gene products | |
| Total | 44 545 253 |
| Electronicb | 43 655 159 |
| Manual | 890 094 |
aMost datasets represent single species; Gramene, the TIGR Gene Index, UniProt GOA and UniProt PDB represent multiple species.
bAnnotations using the IEA (inferred from electronic annotation) evidence code.
New relationship types and new types of links between terms
Initially, GO used two relationship types to link terms: is_a and part_of. The original use of the part_of relationship between regulatory processes and the processes that they regulate did not provide enough specificity to allow users to perform queries that distinguish gene products that play a regulatory role versus a direct role in a biological process. In addition, there were no relationships in the Molecular Function Ontology between regulatory functions and the functions they regulate. In the past two years we have added regulates, positively_regulates, and negatively_regulates relationships between regulatory terms and their regulated parents. The three regulates relationships allow GO to correctly represent important areas of biology where one process affects the manifestation of another process, molecular function, or quality, but may not be a part of that process itself. For example ‘regulation of transcription’ is not a part of ‘transcription’, but lies outside of the transcription process and controls how it unfolds. The regulates relations in GO are used specifically to mean necessarily-regulates, that is: if B regulates A, then whenever B is present, it always regulates A, but A may not always be regulated by B. The introduction of these relationships will allow users to ask important questions about the nature of control processes that underlie much of biology.
Recently, we have also introduced the has_part relationship to GO. It represents a part–whole relationship from the perspective of the parent, and is thus the logical complement to the part_of relationship. In GO, the relationship A has_part B means that A necessarily (always) has B as a part; i.e., if A exists then B also exists as a part of A. If A does not exist, B may or may not exist. For example, ‘cell envelope’ has_part ‘plasma membrane’ means that a cell envelope always has a plasma membrane as a part but a plasma membrane may exist without being a part of a cell envelope.
Perhaps the most significant change is that GO now contains links between its three different branches: MF, BP, and CC. Specifically, there are now part_of relationships between MF and BP and regulates relationships within both MF and BP and between BP and MF. (see examples in Figure 1). A thorough discussion of the various relationship types, both new and old, and their uses in the GO is available at http://geneontology.org/GO.ontology-ext.relations.shtml.
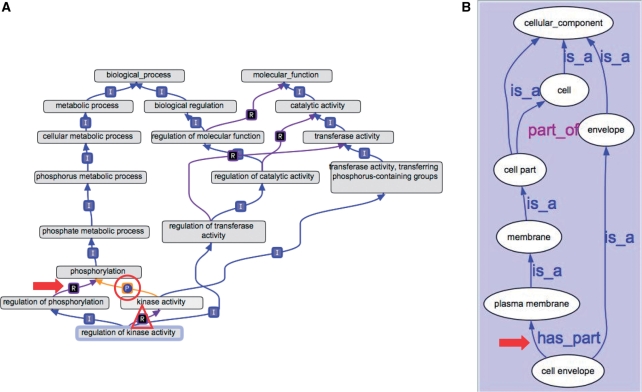
Figure 1.
New intra- and inter-ontology relationships. (A) An example of a regulates link between two BP terms is pointed out by the arrow. An example of a part_of link between an MF term and a BP term is indicated by the circle. A regulates link between a BP term and an MF term is indicated by the triangle. (B) An example of a has_part link between two CC terms is pointed out by the arrow.
The new relationships and new links between ontologies serve several purposes for the user. First, with links between ontologies, annotations can now be propagated from one ontology to another. The most obvious example of this is the propagation of gene-product annotations from a MF term to a BP term when the molecular function has a part_of relationship to a biological process. It is our hope that our users will go beyond this very basic benefit of the cross-ontology links and begin to ask more hypothetical questions using the ontology and annotations to the ontology. For example, a user could now ask what gene products might be involved in regulating a specific metabolic process if they know a regulatory process that controls the metabolic process and they know the types of molecular functions that play roles in the regulatory process.
New ontology files
GO is edited and released on a daily basis. Several versions of GO are available for download (Table 2). An extended version, in OBO 1.2 format, includes the regulates links, the has_part links and the intra-ontology part_of links discussed above and information on when, and by whom, a term was created. Other versions without this additional information are made available to accommodate existing software tools. There are several ways to convert the OBO-format file into the Web Ontology Language (OWL) format (http://www.bioontology.org/wiki/index.php/OboInOwl:Main_Page). These multiple formats allow users to use GO in the ways that they always have, but also give them the opportunity to take advantage of the most recent extensions that the ontology has to offer.
Table 2.
Available GO ontology files
| File name | Content | Format |
|---|---|---|
| gene_ontology_ext.obo | extended | OBO 1.2 |
| gene_ontology.1_2.obo | standard | OBO 1.2 |
| gene_ontology.1_0.obo | standard | OBO 1.0 |
New biological content in the ontology
The GOC continues to add terms and modify the ontology based on annotator and community requests. In addition, by working with community experts and focusing on specific related areas of biology, three areas of GO have seen significant improvements. First, new terms to describe interactions between organisms have been added in collaboration with Plant-Associated Microbe GO (PAMGO) (1). Second, terminology describing the development of branching organs, focusing on the development of the embryonic placenta, prostate gland, salivary gland, mammary gland and lung (2), has been elaborated. Third, the GO branch describing all aspects of heart development has been greatly expanded by the addition of over 200 terms. One of our goals is to provide the research community with the richness of terms required to represent their biology in both breadth and detail. The GOC encourages those who are interested in enriching specific areas of biology in the ontologies to contact and collaborate with us.
Quality control of the ontology and annotations
Among the benefits of the new links and relationship types added to the ontology is that they enable partial automation of ontology-logic quality control. For example, the reasoner built into OBO-Edit can identify missing is_a links in the regulation section of the graph based on existing links in the core biological process graph. Several additional automated checks are performed and corrections are made following curatorial review. We have also begun biological validation of the ontology by comparing annotations of overlapping sets of genes to various cellular GO terms that we expect to be mutually exclusive. Such an analysis reveals potential errors in either annotation or ontology structure. Finally, a check has been designed to identify annotations of gene products from certain species using GO terms that describe processes that do not occur in these species (for example, ‘lactation’ for non-mammalian gene products). We plan to continue to develop both logical and biological checks of the ontology to ensure both its accuracy and completeness.
Sequence Ontology
The Sequence Ontology (SO) (http://sequenceontology.org/) provides the terminology and relations to describe the key features for genomics and other structural sequence annotation. The SO is pioneering the OBO Foundry ontologies in using cross product terms (terms explicitly defined using two or more other ontology terms) to manage ontology editing, with over 194 examples. SO terms are increasingly used in genomic annotation, and have become the standard terminology for annotation sharing and dispersal for the model organism community. There is active development of the ontology and terms and changes can be suggested to SO using the term tracker.
The SO provides tools to both browse the ontology and document it using the miSO browser and the SO wiki. The browser searches either CVS revisions or releases, by term, synonym or ID, and provides a fully reasoned image of the term and relations. The SO wiki provides the user with automated documentation of the status of the term, and the opportunity for manual ‘community based’ documentation. SO also provides tools to support the update of legacy sequence to SO compliant formats such as gff3. Links to all of these resources can be found on the SO web site.
ANNOTATION OF GENE PRODUCTS
GO Consortium members annotate gene products using both electronic and manual methods, refining these methods as new analytical techniques and experimental methods become available. The resulting annotation sets are deposited in the GO repository and GO database and are available for querying using AmiGO. Table 1 shows the current numbers of datasets, annotated gene products, and species in which gene products are annotated.
The Reference Genome project (3) jointly annotates groups of genes across 12 organisms with an emphasis on coordinated annotation of highly conserved genes and genes of particular biomedical importance. Inferences of functions are made using PAINT (Phylogenetic Annotation INferencing Tool) software, which visualizes the annotations in a phylogenetic framework [see Mi et al., this issue (4)]. Reference genome annotation is an important aspect of the GO project in that it takes experimentally derived biological knowledge from a limited number of model organisms and uses that to infer knowledge about similar gene products in other organisms within a phylogenetically based framework. This type of annotation and inference extends the ability of GO to be useful in many different biological contexts.
MAKING GO MORE ACCESSIBLE
OBO-Edit improvements
The GO community develops and uses a freely available Java-based ontology editor, OBO-Edit (5) (http://www.oboedit.org). OBO-Edit 2.0 was released in April 2009 with many improvements to support ontology editors working with updated versions of GO. The new version has completely customizable panel configuration and a graph-based ontology editor as well as improved searching abilities with an auto-complete feature. To support cross products and automated ontology quality control, OBO-Edit 2.0 has enhanced cross product editing, extended reasoning capabilities including a new Rule Based Reasoner, and the ability to assert implied links and remove redundant ones.
AmiGO improvements
AmiGO (6) (http://amigo.geneontology.org), the GO web-based browser, has undergone a large number of improvements with many new features added over several public releases. AmiGO now includes a term enrichment tool (used to find significant shared GO terms or parents of those GO terms in gene products), ontology slimming (used to map annotations of gene products to higher-level terms), community annotation (in association with the GONuts wiki, http://gowiki.tamu.edu), and support for the Reference Genome Project (including special visualizations). AmiGO now displays the regulates relations and includes electronic (IEA) annotations.
Many supporting improvements and changes have been made to AmiGO that improve search quality and the user interface. An ongoing in-place rewrite of the AmiGO code allows for major improvements in graphics, speed, ease of installation, and consistency. Finally, AmiGO now offers search plugins and widgets for all major platforms and the ability for users to try upcoming and experimental software.
Interacting with the user community
GO supports its very active and diverse user community from an email-based helpdesk (http://www.geneontology.org/GO.contacts.shtml). The web-based help documentation has been revised to reflect the new relationship types in the ontology and the new features of the AmiGO browser. GO now communicates news highlights via a dedicated web page, RSS feed and Twitter; these supercede the quarterly newsletter that was emailed to the GO community in previous years.
SUMMARY
The GO Consortium is responsible for the representation of gene product knowledge for a large body of biological data. Over the past several years, we have worked to improve both the logical framework as well as the comprehensiveness of the ontologies. We have now put a system in place that will permit us to continue to extend the representation of biology in GO. New relationships will allow more refined queries to be executed and will begin to allow more hypothesis-generating questions to be asked using the ontology. The improvement of the logical structure will allow for rigorous quality control, ensuring that the ontology is complete and accurate. These improvements will aid in using the ontology for classical gene-clustering experiments by filling in missing relationships that would otherwise result in gene products not being clustered.
Ontology improvement along with continued annotation efforts should make GO an ever more complete representation of the roles that gene products play in a large array of organisms.
FUNDING
National Human Genome Research Institute (NHGRI) (P41 HG02273 to GO PIs J.A.B., M.A., J.M.C., S.L.); GO Consortium member databases receive funding from several National Institutes of Health Institutes [National Human Genome Research Institute (HG000330 to M.G.D., HG02223 to Wormbase, HG004341 to K.E., HG003751 to Reactome, HG01315 to S.G.D., HG002659 to Z.F.I.N.); National Heart, Blood and Lung Institute (HL64541 to R.G.D.), National Institute of General Medical Sciences (U24GM077905, U24GM088849 to EcoliWiki)]; National Science Foundation (DBI# 0703908 to Gramene, DBI#0417062 to TAIR, EF-0523736 to PAMGO); UK Medical Research Council (G0500293 to FlyBase); British Heart Foundation (SP/07/007/23671); European Union Sixth Framework Programme (LSHG-CT-2003-503269 to Reactome). Funding for open access charge: National Human Genome Research Institute (grant #P41 HG02273).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The GO Consortium thanks the community researchers who have participated in content-related meetings or provided valuable feedback on ontology content and annotations.
APPENDIX
Tanya Z. Berardini, Donghui Li, Eva Huala (TAIR, Carnegie Institute for Science, Department of Plant Biology, Stanford, CA, USA); Susan Bridges, Shane Burgess, Fiona McCarthy (AgBase, Mississippi State University; MS, USA); Seth Carbon, Suzanna E. Lewis, Christopher J. Mungall, Amina Abdulla (BBOP, LBNL, Berkeley, CA, USA); Valerie Wood (Cancer Research UK, London UK); Erika Feltrin, Giorgio Valle (CRIBI, University of Padua, Italy); Rex L. Chisholm, Petra Fey, Pascale Gaudet, Warren Kibbe, Siddhartha Basu, Yulia Bushmanova (dictyBase, Northwestern University, Chicago, IL, USA); Karen Eilbeck (Eccles Institute of Human Genetics, University of Utah, Salt Lake City, UT, USA); Deborah A. Siegele, Brenley McIntosh, Daniel Renfro, Adrienne Zweifel and James C. Hu (EcoliWiki, Departments of Biology and Biochemistry and Biophysics, Texas A&M Univ., College Station, TX, USA); Michael Ashburner, Susan Tweedie (FlyBase, Department of Genetics, University of Cambridge, Cambridge, UK); Yasmin Alam-Faruque, Rolf Apweiler, Andrea Auchinchloss, Amos Bairoch, Daniel Barrell, David Binns, Marie-Claude Blatter, Lydie Bougueleret, Emmanuel Boutet, Lionel Breuza, Alan Bridge, Paul Browne, Wei Mun Chan, Elizabeth Coudert, Louise Daugherty, Emily Dimmer, Ruth Eberhardt, Anne Estreicher, Livia Famiglietti, Serenella Ferro-Rojas, Marc Feuermann, Rebecca Foulger, Nadine Gruaz-Gumowski, Ursula Hinz, Rachael Huntley, Silvia Jimenez, Florence Jungo, Guillaume Keller, Kati Laiho, Duncan Legge, Philippe Lemercier, Damien Lieberherr, Michele Magrane, Claire O'D;onovan, Ivo Pedruzzi, Sylvain Poux, Catherine Rivoire, Bernd Roechert, Tony Sawford, Michel Schneider, Eleanor Stanley, Andre Stutz, Shyamala Sundaram, Michael Tognolli, Ioannis Xenarios (GOA-UniProtKB: EBI, Hinxton, UK and SIB, Geneva, Switzerland); Midori A. Harris, Jennifer I. Deegan (née Clark), Amelia Ireland, Jane Lomax (GO-EBI, Hinxton, UK); Pankaj Jaiswal (Gramene, Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR, USA); Marcus Chibucos, Michelle Gwinn Giglio, Jennifer Wortman (Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, MD, USA); Linda Hannick, Ramana Madupu (The J. Craig Venter Institute, Rockville, MD, USA); David Botstein, Kara Dolinski, Michael S. Livstone, Rose Oughtred (Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ, USA); Judith A. Blake, Carol Bult, Alexander D. Diehl, Mary Dolan, Harold Drabkin, Janan T. Eppig, David P. Hill, Li Ni, Martin Ringwald, Dmitry Sitnikov (MGI, The Jackson Laboratory, Bar Harbor, ME, USA); Candace Collmer (PAMGO, Wells College, Aurora, NY, USA); Trudy Torto-Alalibo (PAMGO, Virginia Bioinformatics Institute, VA, USA); Stan Laulederkind, Mary Shimoyama, Simon Twigger (RGD, Medical College of Wisconsin, Milwaukee, WI, USA); Peter D’Eustachio, Lisa Matthews (Reactome, Department of Biochemistry, NYU School of Medicine, New York NY USA); Rama Balakrishnan, Gail Binkley, J. Michael Cherry, Karen R. Christie, Maria C. Costanzo, Stacia R. Engel, Dianna G. Fisk, Jodi E. Hirschman, Benjamin C. Hitz, Eurie L. Hong, Cynthia J. Krieger, Stuart R. Miyasato, Robert S. Nash, Julie Park, Marek S. Skrzypek, Shuai Weng, Edith D. Wong (SGD, Department of Genetics, Stanford University, Stanford, CA, USA); Martin Aslett (Wellcome Trust Sanger Institute, Hinxton, UK); Juancarlos Chan, Ranjana Kishore, Paul Sternberg, Kimberly Van Auken (WormBase, California Institute of Technology, Pasadena, CA, USA); Varsha K. Khodiyar, Ruth C. Lovering, Philippa J. Talmud (UCL, London, UK); Doug Howe, Monte Westerfield (ZFIN, University of Oregon, Eugene, OR, USA).
REFERENCES
- 1.Torto-Alalibo T, Collmer CW, Gwinn-Giglio M. The Plant-Associated Microbe Gene Ontology (PAMGO) Consortium: community development of new Gene Ontology terms describing biological processes involved in microbe-host interactions. BMC Microbiol. 2009;9(Suppl. 1):S1. doi: 10.1186/1471-2180-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill DP, Sitnikov D, Blake JA. Using gene ontology to study branching morphogenesis in mice. Dev. Biol. 2009;331:454. [Google Scholar]
- 3.The Reference Genome Group of the Gene Ontology Consortium. The Gene Ontology's; Reference Genome Project: a unified framework for functional annotation across species. PLoS Comput. Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs, and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2009;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Day-Richter J, Harris MA, Haendel M, Lewis S. OBO-Edit—an ontology editor for biologists. Bioinformatics. 2007;23:2198–2200. doi: 10.1093/bioinformatics/btm112. [DOI] [PubMed] [Google Scholar]
- 6.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]