Abstract
The identification of orthologous relationships forms the basis for most comparative genomics studies. Here, we present the second version of the eggNOG database, which contains orthologous groups (OGs) constructed through identification of reciprocal best BLAST matches and triangular linkage clustering. We applied this procedure to 630 complete genomes (529 bacteria, 46 archaea and 55 eukaryotes), which is a 2-fold increase relative to the previous version. The pipeline yielded 224 847 OGs, including 9724 extended versions of the original COG and KOG. We computed OGs for different levels of the tree of life; in addition to the species groups included in our first release (i.e. fungi, metazoa, insects, vertebrates and mammals), we have now constructed OGs for archaea, fishes, rodents and primates. We automatically annotate the non-supervised orthologous groups (NOGs) with functional descriptions, protein domains, and functional categories as defined initially for the COG/KOG database. In-depth analysis is facilitated by precomputed high-quality multiple sequence alignments and maximum-likelihood trees for each of the available OGs. Altogether, eggNOG covers 2 242 035 proteins (built from 2 590 259 proteins) and provides a broad functional description for at least 1 966 709 (88%) of them. Users can access the complete set of orthologous groups via a web interface at: http://eggnog.embl.de.
INTRODUCTION
Next-generation sequencing technologies are now generating a vast amount of sequence data. This leads to a dramatic increase in the number of predicted protein sequences, which serve as a starting point for structural, functional and phylogenomic studies. In such studies, high-throughput comparative analyses are often required to transfer information between organisms, for which the concept of orthology is crucial. The original definition by Fitch (1) describes orthologs as genes that diverged through a speciation event, as opposed to paralogs, which diverged after a duplication event. This has been extended and refined by introducing the concepts of orthologous groups (OGs) (2), in-paralogs and out-paralogs (3,4). In practice, however, the identification and classification of homologous genes remain very difficult and rely on operational definitions. An enormous effort is being put into the development of different approaches to establish orthologous relationships between genes from different genomes. This includes several algorithms using the simple graph-based methods, including reciprocal-best-hit approach (5), identification of best-hit triangles (2,6–8) and clustering-based approaches (9–11) as well as tree-based methods (12–16).
In addition to the quality of the grouping of genes, the practical usability of OGs is determined by the ability to provide a robust functional annotation. Thus, newer projects not only aggregate orthology information from various sources to allow comparison between methods but also aim to provide annotation tools (17,18). Nevertheless, evolutionary genealogy of genes: non-supervised OGs (eggNOG) (19) and the COG/KOG/arCOG resources (2,6,7) are still the only databases providing explicit functional annotations for the OGs at different hierarchical levels, whereby the COG/KOG resource is based on a robust manual expert annotation, which eggNOG is using and automatically extending (19).
Here, we describe the new features of the second version of eggNOG, a resource that provides OGs from the three domains of life at several levels of resolution. eggNOG v2 contains twice as many species and proteins as the previous version, additional hierarchical levels allowing higher resolution for a number of taxonomic groups, new annotation sources and an extended interface for an in-depth analysis of orthologous relationships.
CONSTRUCTION OF HIERARCHICAL OGs
The automated procedure described previously (19) has been used to assemble proteins into OGs from 630 complete genomes (529 bacteria, 46 archaea and 55 eukaryotes). Complete proteomes were downloaded from the RefSeq (20), Ensembl (21), GiardiaDB (22) or TAIR (23) databases. This particular data set also forms the basis for STRING v8 (24) and STITCH v2 (25), allowing for easy integration across these databases.
Altogether, the protein data set covers 2 590 259 proteins of which 2 242 035 (87%) were included in at least one of 224 847 OGs generated by eggNOG. The growing number of species and proteins included in this release drastically increased the computational time. All-against-all similarity searches have therefore been performed using Basic Local Alignment Search Tool (BLAST) (26) instead of the Smith–Waterman algorithm (27).
Compared to the 4873 COGs and the 4850 KOGs that are constructed across all three domains of life and for all eukaryotes, respectively, this procedure assembles additional proteins into NOGs (440 359 proteins into 59 497 NOGs and 181 427 into 17 845 euNOGs). These complement the published COGs and KOGs built respectively for 66 and seven species (6), which are extended in eggNOG to cover 630 species encompassing, respectively, 1 547 381 and 483 043 proteins.
To provide a higher resolution of OGs in frequently used taxonomic groupings, we applied our procedure to several subsets of organisms separately. We updated the previously computed more fine-grained NOGs at the level of fungi (fuNOGs), metazoans (meNOGs), insects (inNOGs), vertebrates (veNOGs) and mammals (maNOGs) and added groups for archaea (arNOGs), fishes (fiNOGs), rodents (roNOGs) and primates (prNOGs).
Extending the automated annotation of protein function
An important feature of eggNOG is the functional annotations of the OGs. Our original pipeline, providing functional descriptions for the NOGs, is now complemented by an automatic inference of functional categories (FCs) which were taken from the COG database (2). The 25 FCs available from the COG resource have been widely used to assess comparative genomics studies and will enable higher-order analyses of OGs identified in any data set.
We use two complementary methods to infer FCs of OGs based on the 4617 COGs (used for NOGs and arNOGs) and 4381 KOGs (used for all other OGs). The first method uses Support Vector Machines (SVM) trained on the COGs and KOGs to classify NOGs into the 25 FCs based on feature vectors. Two feature vectors were created for each OG. One was built from functional information mapped onto the eggNOG protein data set, including KEGG pathways and modules (28), GO terms (29), SMART domains (30), PFAM domains (31), UniProt keywords (32) and words from UniProt/RefSeq (20) description lines. The second feature vector includes also words from MEDLINE abstracts referring to a particular protein (24). Each attribute in the feature vector encodes the fraction of proteins in the group having the feature in question.
The second method for assigning FCs makes use of the hierarchical structure of eggNOG, namely that the same proteins can be assigned to OGs at several levels in the tree of life (e.g. a KOG and a meNOG). In case an FC could not be assigned to a NOG by the SVM method, we check if most of the proteins in the NOG belong to a common functionally annotated COG or KOG, in which case we transfer the FCs from the coarse-grained level (COGs or KOGs) to the more fine-grained one (e.g. arNOGs or meNOGs). The assignment of an FC to a single NOG is achieved on the basis of a coverage value determined by the occurrence of that FC (via the proteins shared with the reference level) in respect to the total number of proteins in that NOG.
ANNOTATION RESULTS
In addition to providing functional annotations via description lines for many NOGs (19), we are now able to predict functional categories as well. At the universal level, our function annotation pipeline provides description lines for 14 956 (25%) and an FC for 6262 (11%) of the 59 497 coarse-grained NOGs. At the eukaryotic level, 7566 euNOGs (52%) have a description line and 4120 (34%) have an FC. In addition, eggNOG contains 137 782 more fine-grained OGs of which 100 750 (73%) and 89 232 (65%) have been annotated with a description line and an FC, respectively (Table 1).
Table 1.
Annotation statistics at different taxonomic levels
| Level | OG count | Description line |
Functional categories |
||
|---|---|---|---|---|---|
| Annotated | (%) | Annotated | (%) | ||
| COG + NOG | 64 370 | 4474 + 14 956 | 30.2 | 2824 + 6262 | 14.1 |
| arNOG | 9809 | 4144 | 42.2 | 4540 | 46.3 |
| KOG + euNOG | 22 695 | 4288 + 7566 | 52.2 | 3514 + 4120 | 33.6 |
| fuNOG | 9976 | 5661 | 56.7 | 5775 | 57.9 |
| meNOG | 22 691 | 16 636 | 73.3 | 13 490 | 59.5 |
| inNOG | 8049 | 5034 | 62.5 | 5810 | 72.2 |
| veNOG | 21 357 | 16 722 | 78.3 | 13 291 | 62.2 |
| fiNOG | 13 674 | 8903 | 65.1 | 9580 | 70.1 |
| maNOG | 20 222 | 16 959 | 83.9 | 13 075 | 64.7 |
| roNOG | 14 038 | 11 918 | 84.9 | 10 547 | 75.1 |
| prNOG | 17 966 | 14 773 | 82.2 | 13 124 | 73.0 |
At the levels for COGs (universal) and KOGs (eukaryotes) the additional automatically generated non-supervised orthologous groups NOGs and euNOGs, respectively, are separated.
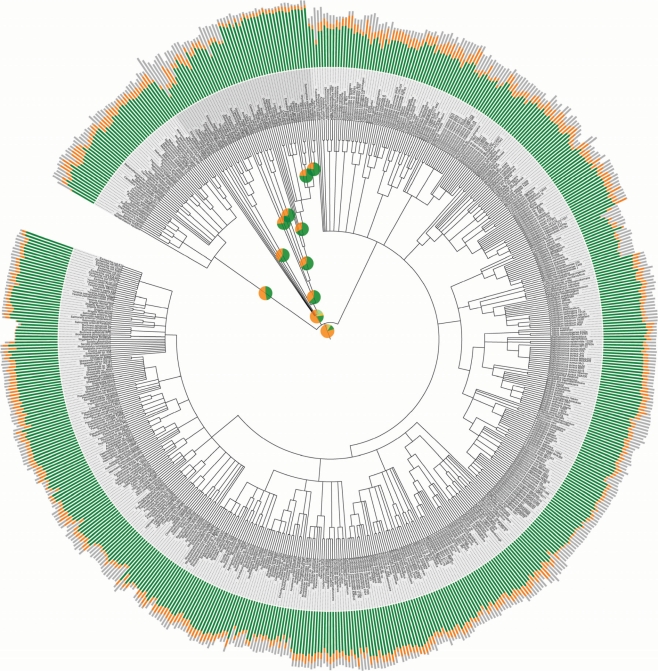
This enables us to assign 2 242 035 of the 2 590 259 genes (87% of the genes in the analyzed genomes) to an OG and to provide at least a broad functional description or FC for 1 966 709 of them (78% of the genes that could be assigned to an OG). The corresponding numbers for each set of OGs as well as for each individual genome are summarized in Figure 1.
Figure 1.
Statistics on the content of the eggNOG database. The eggNOG assignments for 630 complete genomes were mapped onto the tree of life. The stacked bar charts outside the tree show the proportion of genes from each genome that can be assigned to a functionally annotated orthologous group (green), an unannotated orthologous group (orange) or no orthologous group (gray). The length of each bar is proportional to the logarithm of the number of genes in the respective genome. The pie charts inside the tree show the fractions of orthologous groups at each level in the hierarchy that could be annotated with a functional category (green for NOGs, light green for extended COGs and KOGs) or not (orange for NOGs, light orange for extended COGs and KOGs). An interactive version is available in the ‘Overview’ section at: http://eggnog.embl.de. This figure was made using iTOL.
Extended features in eggNOG v2.0
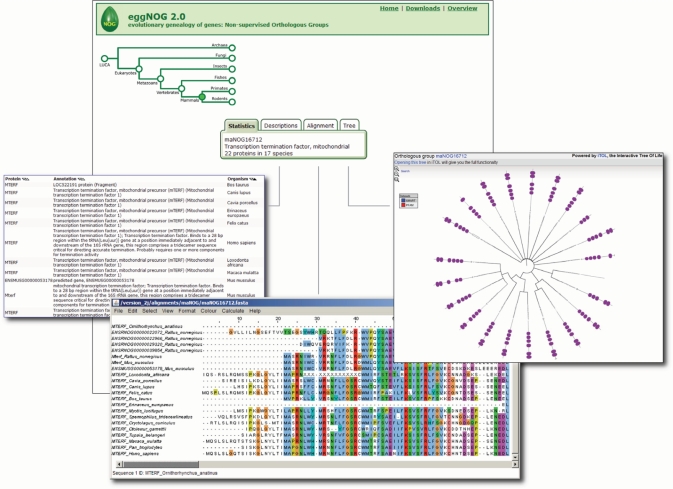
To facilitate the in-depth analysis of the orthologous relationships within the groups of proteins, we now provide precomputed high-quality Multiple Sequence Alignments (MSAs) and maximum-likelihood trees via the web interface (Figure 2).
Figure 2.
Screenshot of the detailed results page. The eggNOG database was queried for the term ‘mTERF’, the mitochondrial precursor of the transcription termination factor 1. The navigation tree at the top of the page allows the user to change the view to more coarse-grained orthologous groups, for example, the mammalian orthologous groups. The tab menu, shown here, enables several in-depth interactions with the new data (i.e. MSA or phylogenetic trees, here displayed with SMART domains).
Numerous methods are available to build MSAs [e.g. ClustalW (33), Muscle (34), MAFFT (35) and PRANK (36)] but some programs appear to be more suitable for particular protein families than others (37). Thus, we applied a new approach, named Automated QUality improvement for multiple sequence Alignments (AQUA) (Muller et al., submitted for publication), which combines existing tools to deliver high-quality MSAs.
The construction of the different phylogenetic trees was carried out using the following steps. One hundred bootstrap replicates were created from the MSA using the SEQBOOT program from the Phylip package (38). Following this, PhyML (39) was used to find the maximum-likelihood tree for each of the 100 bootstrap replicates and for the original alignment using default parameters. Finally, a consensus tree was constructed, using the CONSENSE program from the Phylip package. We used ReadSeq (40) to convert between the different sequence file formats used by those programs.
ACCESS OPTIONS
The eggNOG resource can be queried via a web interface; data can be downloaded under the Creative Commons Attribution 3.0 License at: http://eggnog.embl.de or via FTP at: ftp://eggnog.embl.de/eggNOG/2.0/. Gene and protein names, database identifiers, amino acid sequences, or OG names can be used to query the database. As a default, the most fine-grained OGs available are displayed for maximal resolution. The user can navigate among the different levels of orthology using an available guide-tree of organisms to find the desired balance between phylogenetic coverage and functional specificity within our hierarchy of OGs. Through the new interface, users can access different information panels encompassing the detailed list of proteins belonging to a particular OG as well as the corresponding MSA and phylogenetic tree. The MSA can be interactively displayed using the Jalview applet (41) or downloaded in FASTA format. The phylogenetic trees are accessed through a dedicated iTOL (42) viewer together with mapped PFAM and SMART domains, via the ATV program applet (43), or can be downloaded in Newick format.
CONCLUSIONS/PERSPECTIVES
With 630 genomes covered, an increased OG hierarchy, and a high coverage of newly categorized functional annotation, the new version of eggNOG is one of the most comprehensive and complete resources for deciphering the orthologous relationships between proteins from various species. The changes and improvements in the interface and the availability of the OGs for download will not only facilitate the daily use of the database, but also the integration of eggNOG in high-throughput comparative genomics studies. Our future plans include the addition of more complete genomes and development of a more scalable and flexible pipeline for generating the groups.
FUNDING
EMBL, the European Commission Programme, Eurasnet EU [Grant LSHG-CT-2005-518238 (FP6), IMPACT 213037 (FP7)]; the Novo Nordisk Foundation Center for Protein Research, the Swiss Institute of Bioinformatics; and the University of Zurich (partial, through its Research Priority Program ‘Systems Biology and Functional Genomics’). This work was supported in part by the Bundesministerium fuer Bildung und Forschung (Nationales Genomforschungsnetz Foerderkennzeichen 01GS08169). Funding for open access charge: European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 2.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 4.Sonnhammer EL, Koonin EV. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 5.Berglund AC, Sjolund E, Ostlund G, Sonnhammer EL. InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res. 2008;36:D263–D266. doi: 10.1093/nar/gkm1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol. Direct. 2007;2:33. doi: 10.1186/1745-6150-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kriventseva EV, Rahman N, Espinosa O, Zdobnov EM. OrthoDB: the hierarchical catalog of eukaryotic orthologs. Nucleic Acids Res. 2008;36:D271–D275. doi: 10.1093/nar/gkm845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth AC, Gonnet GH, Dessimoz C. Algorithm of OMA for large-scale orthology inference. BMC Bioinformatics. 2008;9:518. doi: 10.1186/1471-2105-9-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchiyama I. MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 2007:D343–D346. doi: 10.1093/nar/gkl978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Heijden RT, Snel B, van Noort V, Huynen MA. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinformatics. 2007;8:83. doi: 10.1186/1471-2105-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wapinski I, Pfeffer A, Friedman N, Regev A. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics. 2007;23:i549–i558. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- 14.Huerta-Cepas J, Bueno A, Dopazo J, Gabaldon T. PhylomeDB: a database for genome-wide collections of gene phylogenies. Nucleic Acids Res. 2008;36:D491–D496. doi: 10.1093/nar/gkm899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta RS, Meacham C, Samad B, Neyer C, Sjolander K. Berkeley PHOG: PhyloFacts orthology group prediction web server. Nucleic Acids Res. 2009:W84–W89. doi: 10.1093/nar/gkp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyre TA, Wright MW, Lush MJ, Bruford EA. HCOP: a searchable database of human orthology predictions. Brief Bioinform. 2007;8:2–5. doi: 10.1093/bib/bbl030. [DOI] [PubMed] [Google Scholar]
- 18.Kuzniar A, Lin K, He Y, Nijveen H, Pongor S, Leunissen JA. ProGMap: an integrated annotation resource for protein orthology. Nucleic Acids Res. 2009;37:W428–W434. doi: 10.1093/nar/gkp462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aurrecoechea C, Brestelli J, Brunk BP, Carlton JM, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, et al. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 2009;37:D526–D530. doi: 10.1093/nar/gkn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saebo PE, Andersen SM, Myrseth J, Laerdahl JK, Rognes T. PARALIGN: rapid and sensitive sequence similarity searches powered by parallel computing technology. Nucleic Acids Res. 2005;33:W535–W539. doi: 10.1093/nar/gki423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Universal Protein Resource (UniProt) Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 36.Loytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320:1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JD, Koehl P, Ripp R, Poch O. BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 38.Felsenstein J. PHYLIP – Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 39.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert D. Sequence file format conversion with command-line readseq. Curr. Protoc. Bioinformatics. 2003 doi: 10.1002/0471250953.bia01es00. Appendix 1, Appendix 1E. [DOI] [PubMed] [Google Scholar]
- 41.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 43.Zmasek CM, Eddy SR. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]