Abstract
The Plant Transcription Factor Database (PlnTFDB; http://plntfdb.bio.uni-potsdam.de/v3.0/) is an integrative database that provides putatively complete sets of transcription factors (TFs) and other transcriptional regulators (TRs) in plant species (sensu lato) whose genomes have been completely sequenced and annotated. The complete sets of 84 families of TFs and TRs from 19 species ranging from unicellular red and green algae to angiosperms are included in PlnTFDB, representing >1.6 billion years of evolution of gene regulatory networks. For each gene family, a basic description is provided that is complemented by literature references, and multiple sequence alignments of protein domains. TF or TR gene entries include information of expressed sequence tags, 3D protein structures of homologous proteins, domain architecture and cross-links to other computational resources online. Moreover, the different species in PlnTFDB are linked to each other by means of orthologous genes facilitating cross-species comparisons.
INTRODUCTION
In order to fulfil their biological functions, genes must be expressed in specific spatiotemporal patterns. These patterns are to a large extent established by controlling the transcription of the genes through which RNA copies are generated from the DNA template. In this process, a protein complex composed of general transcription factors (TFs) is mandatory to sustain the expression of all genes encoded by the genome. In addition, other regulatory proteins enhance or repress the transcriptional rate of target genes in response to biotic and abiotic stimuli, and intrinsic developmental processes. These proteins are TFs that bind, in a sequence-specific manner, to cis-elements in the target promoters, and other transcriptional regulators (TRs) that exert their regulatory function through protein–protein interactions or chromatin remodeling. The identification of such TFs and TRs from an appreciable number of organisms of divergent lineages represents an important first step towards the understanding of gene regulatory networks and their evolution. For plants, this step has already been made by several groups through the development of databases dedicated to the presentation of TFs and TRs and accompanying information of relevance to the research community (1–6). Here we present the current status of the Plant Transcription Factor Database, PlnTFDB (4), which in its updated version (v3.0) provides information about the putatively complete sets of TFs and TRs from 19 plant species (sensu lato) encompassing a broad phylogenetic range of >1.6 billion years of divergent evolution (7).
DATA SOURCES, ANALYSES AND IMPLEMENTATION
Species and proteomes covered
In order to identify putatively complete sets of TFs and TRs, we applied our previously established analysis pipeline to the proteomes of species whose genomes have been completely sequenced and annotated (4). The PlnTFDB v3.0 covers 19 different plant species ranging from unicellular red and green algae to angiosperms, therewith expanding the species spectrum of the previous version by 12 new species. The species analysed and the sources of the sequence data used to establish PlnTFDB v3.0 are listed in Table 1.
Table 1.
Species analysed and number of families and classified proteins per species
| Groups | Species | Source | Annotation version | Reference | Total number of proteinsa | Genome size (Mbp) | Number of families | Number of classified proteinsa |
|---|---|---|---|---|---|---|---|---|
| Red algae (Rhodophytes) | Cyanidioschyzon merolae | 1 | 20070710 | (8) | 5008 | 16.52 | 34 | 147 |
| Galdieria sulphuraria | 9 | (9) | 6604 | 10 | 37 | 201 | ||
| Green algae (Prasinophytes) | Micromonas pusilla CCMP1545 | 2 | 2 | (10) | 10 455 | 15 | 49 | 289 |
| Micromonas sp. RCC299 | 2 | 3 | (10) | 10 160 | 15 | 49 | 326 | |
| Ostreococcus tauri | 2 | 2 | (11) | 7812 | 12.56 | 47 | 216 | |
| Ostreococcus lucimarinus | 2 | 2 | (11) | 7651 | 13.204 | 46 | 236 | |
| Green algae (Chlorophytes) | Chlamydomonas reinhardtii | 2 | 4 | (12) | 16 460 | 121 | 52 | 346 |
| Chlorella sp. NC64A | 2 | 1 | 9762 | 40 | 48 | 304 | ||
| Coccomyxa sp. C-169 | 2 | 1 | 10 174 | 120 | 47 | 261 | ||
| Bryophyte (Bryopsida) | Physcomitrella patens | 2 | 1.1 | (13) | 35 724 | 480 | 72 | 1295 |
| Spike-moss (Lycopodiophyte) | Selaginella moellendorffii | 2 | 1 | 22 138 | 100 | 74 | 896 | |
| Angiosperms (Monocots) | Oryza sativa subsp. indica | 3 | 20050118 | (14) | 49 643 | 420 | 79 | 2393 |
| Oryza sativa subsp. japonica | 4 | 6 | (15) | 63 306 | 420 | 79 | 2722 | |
| Sorghum bicolor | 2 | 4 | (16) | 35 682 | 730 | 78 | 2231 | |
| Zea mays | 5 | 3b.50 | 55 810 | 2400 | 79 | 3608 | ||
| Angiosperms (Eudicots) | Carica papaya | 7 | (17) | 24 852 | 372 | 81 | 1480 | |
| Arabidopsis lyrata | 2 | 1 | 32 234 | 206.7 | 81 | 2162 | ||
| Arabidopsis thaliana | 6 | 8 | (18) | 30 707 | 125 | 81 | 2451 | |
| Populus trichocarpa | 2 | 1.1 | (19) | 45 009 | 485 | 81 | 2901 | |
| Vitis vinifera | 8 | 1 | (20) | 30 342 | 500 | 80 | 1725 |
(1) CME GP, Cyanidioschyzon merolae Genome Project, http://merolae.biol.s.u-tokyo.ac.jp/; (2) JGI/DOE, Joint Genome Institute/Department of Energy, http://www.jgi.doe.gov/; (3) BGI, Beijing Genomics Institute, http://www.genomics.org.cn/; (4) TIGR, The Institute for Genomic Research, http://www.tigr.org/; (5) MaizeSequence.org, http://www.maizesequence.org; (6) TAIR, The Arabidopsis Information Resource, http://www.arabidopsis.org/; (7) The Hawaii Papaya Genome Project, http://asgpb.mhpcc.hawaii.edu/papaya/; (8) Genoscope, Centre Nacional de Séquençage http://www.genoscope.cns.fr/spip/Vitis-vinifera-e.html; (9) Data communicated by Prof. Dr Andreas Weber, University of Duesseldorf, Germany.
aNumber of non-redundant proteins.
Identification of protein domains and new domain models
The identification of TFs and TRs and their classification into families exploits the presence of protein domains and their combination within proteins (4). To generate the current release of PlnTFDB, domains were identified using the Pfam protein families database v23.0 (21) and the software package HMMER v2.3.2 (http://hmmer.janelia.org/). Domain hits with a score higher than or equal to the gathering cut-off (–cut_ga) defined for each hidden Markov model (HMM) were kept for further analyses.
For some families, there is no domain represented in the Pfam database; in such cases we developed profile HMMs based on sequence alignments of the respective domains. For the current version of PlnTFDB, we established HMMs for the characteristic domains of the families NOZZLE and VARL. An HMM for the NOZZLE family is available in the Pfam database; however, this model only recovers members from the Brassicaceae family (e.g. Arabidopsis sp.). Hence we used the Arabidopsis thaliana sequences to perform a PSI-BLAST search against the non-redundant protein database at NCBI (http://www.ncbi.nlm.nih.gov/). This allowed us building a multiple sequence alignment and HMM of NOZZLE proteins from several angiosperms, i.e. A. thaliana, Brassica juncea, Medicago truncatula and Vitis vinifera.
The HMM for the VARL family was built by using the alignment reported in Duncan et al. (22), with sequences from Chlamydomonas reinhardtii and Volvox carteri. The alignments used to create the new HMMs are available through the database web interface.
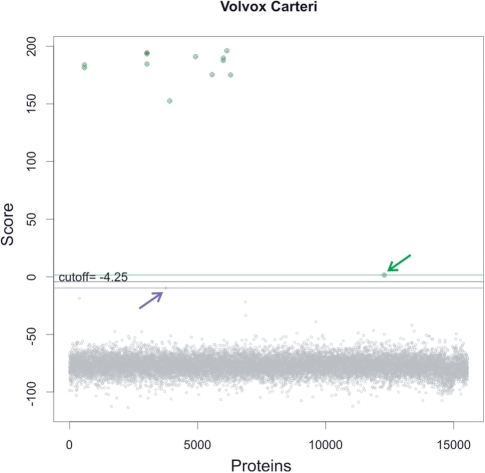
After building these HMMs, a score threshold had to be defined, beyond which the hits are considered significant. To this end, we run an HMM search with the newly created models using a very permissive preliminary threshold (e-value ≤ 10). Subsequently, the known members of the family were localized within the list of hits, which allowed us identifying putative true positives (TPs) and putative true negatives (TNs), thus defining the score threshold as the average between the minimum score obtained by a TP and the maximum score obtained by a TN. This procedure is illustrated in Figure 1.
Figure 1.
Selecting the significance score threshold in newly created profile HMMs. The graphic shows the scores obtained for proteins in the V. carteri proteome when searched with the VARL HMM with an e-value cut-off of 10. Known members of the family in this species (TPs) are highlighted in green. The putative TN with the highest score is indicated by a purple arrow. The TP with the minimum score is highlighted by a green arrow. The significance score threshold (black line) is computed as the average between the minimum score for TPs (green line) and the maximum score for TNs (purple line). For this family, the selected threshold is −4.25 bits.
Rules for the classification of TFs and TRs
Compared with version 2.0 of the database, we have increased the number of rules established for the classification of TFs and TRs by Riaño-Pachón et al. (4). We have now included 16 additional families, totalling 84 in PlnTFDB v3.0. Briefly, the classification rules ask for the presence of a single domain in 77 cases, and a combination of domains in the remaining 7 cases. In addition to these ‘required’ domains, the rules for some families include ‘forbidden’ domains. The forbidden domains allow establishing a mutually exclusive classification system ensuring that each individual protein is classified as a member of a single TF or TR family only. The current sets of ‘required’ and ‘forbidden’ domains of each individual family are listed in Supplementary Data, Appendix 1. We included two meta-rules in our classification scheme: (i) if a protein harbours domains characteristic of a TF family and a TR family, we assigned it to the TF family, e.g. A. thaliana protein AT3G51120.1 could be assigned to families C3H (TF) and SWI/SNF-BAF60b (TR), but according to this meta-rule it is assigned to C3H. (ii) When the protein of interest contains domains characteristic of more than one TF family or more than one TR family, it was assigned to the family to which its characteristic domains matched with the lowest e-value. For example, protein 425147 from Selaginella moellendorffii could be classified as C2H2 (TF, e-value 7.3e-3) or RWP-RK (TF, e-value 6.1e-11), according to the meta-rule it was assigned to the RWP-RK family.
Database interface and availability
The information about the different regulatory proteins and their classification into families, as well as sequence alignments, 3D structures, literature references and links to other databases are stored in a relational database, powered by MySQL (http://www.mysql.com; database schema in Supplementary Data, Appendix 2). The interface of the database to the World Wide Web (WWW) was developed by using PHP, JavaScript and Java applets (Jmol, http://www.jmol.org/; and Jalview, http://www.jalview.org/) following HTML 4.01 and CCS v2.1 W3 standards to ensure browser interoperability.
PlnTFDB can be queried using keywords or sequences (using blastp or blastx), and it is freely accessible through the WWW via http://plntfdb.bio.uni-potsdam.de/v3.0/ using any modern web browser. The Java Runtime Environment (JRE) 1.6.0.12 or newer is required in order to visualize domain alignments and protein 3D structures.
3D PROTEIN STRUCTURES, EXPRESSED SEQUENCE TAGS AND ORTHOLOGUES
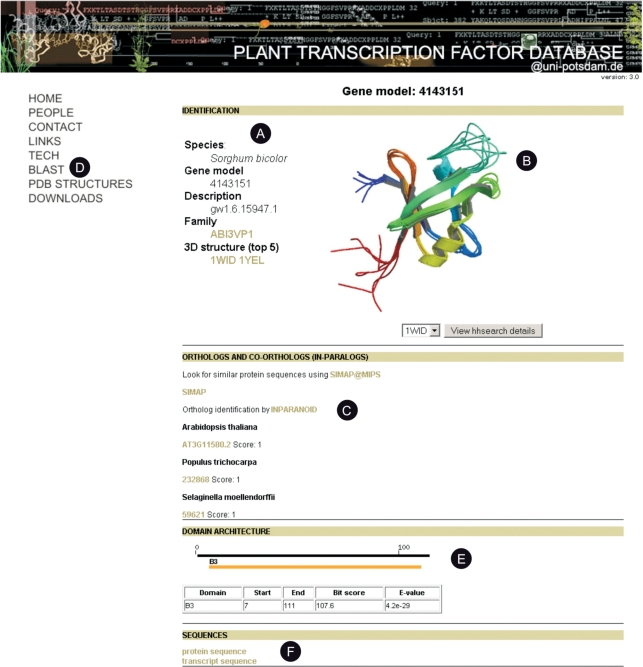
To widen the information provided for each TF and TR in PlnTFDB, we have performed similarity-based searches against the database of sequences with known protein tertiary structures available from the Protein Data Bank (PDB) and the expressed sequence tag (EST) databases available from GenBank. To identify related ESTs, we used BLAST as search engine, keeping as significant all hits with an e-value ≤ 10−10 and an alignment identity of ≥50% over a length of ≥80 amino acids. For the detection of homologous 3D protein structures, we used the package hhsearch (http://toolkit.tuebingen.mpg.de/hhpred) that employs HMM—HMM comparisons to detect remote homologues. Hits were considered significant if the probability of the target being a TP was >98%. The 3D structures of proteins similar to entries in PlnTFDB can be visualized with the Jmol applet (Figure 2), and links are provided to the PDB web site.
Figure 2.
Screenshot of a web page displaying details for a TF gene in PlnTFDB. (A) Every gene page in PlnTFDB displays basic information (including species name and gene family assignment) for a given TF or TR. If gene names had been assigned (only for A. thaliana and O. sativa ssp. japonica) they will be displayed as well. (B) The best hits (hhsearch, probability of being a TP ≥98%) to PDB protein 3D structures are visualized as static images, a link is provided to the embedded Java applet Jmol where basic operations on the 3D structure can be performed. (C) Links to orthologues in PlnTFDB are provided. (D) Users can query PlnTFDB through similarity searches (BLAST) using a protein or a nucleotide sequence as query. (E) Domain architecture is displayed with links to the original domain databases (Pfam or our local database, see section ‘Identification of protein domains and new domains models’). (F) Links to the protein and transcript sequences of the gene are provided.
The genomes of some species covered by PlnTFDB, e.g. A. thaliana and Oryza sativa ssp. japonica, are relatively well annotated with respect to the biological functions of the proteins they encode, whereas genomes of others, including C. reinhardtii, are still in a preliminary status of annotation of biological functions. As orthologous genes often have the same function in different species (23), we have used InParanoid (24) to detect clusters of orthologous genes between pairs of species in PlnTFDB. This will ease the transfer of functional information and provide effective cross-references among the species in PlnTFDB.
QUALITY CONTROL
To evaluate the quality of the putatively complete sets of TFs and TRs reported in PlnTFDB, we compared our predictions to published datasets on detailed single-family phylogenetic studies, and defined the published analyses as gold standards. We calculated the sensitivity and the positive predicted value (PPV) as described before (4). The results of this evaluation are shown in Table 2. In all cases, both measures are >80%, and for most families the sensitivity and PPV values are >90% (shown in bold face in Table 2), evidencing low rates of false negatives (FNs) and positives (FPs).
Table 2.
Sensitivity and PPV of PlnTFDB predictions
| Species | Family | Reference | TP/TP + FN | TP/TP + FP | Sensitivity | PPV |
|---|---|---|---|---|---|---|
| ATH | AP2-EREBP | (25) | 146/147 | 146/146 | 0.99 | 1.00 |
| ARF | (26) | 21/23 | 21/23 | 0.91 | 0.91 | |
| AUX/IAA | (26) | 28/29 | 28/28 | 0.97 | 1.00 | |
| bHLH | (27) | 125/154 | 125/136 | 0.81 | 0.92 | |
| bZIP | (28) | 70/76 | 70/70 | 0.92 | 1.00 | |
| C2C2-Dof | (29) | 35/36 | 35/36 | 0.97 | 0.97 | |
| C2C2-GATA | (30) | 29/29 | 29/29 | 1.00 | 1.00 | |
| C3H | (31) | 65/67 | 65/68 | 0.97 | 0.96 | |
| GRAS | (32) | 32/32 | 32/33 | 1.00 | 0.97 | |
| MADS | (33) | 97/105 | 97/105 | 0.92 | 0.92 | |
| MADS | (34) | 98/108 | 98/105 | 0.91 | 0.93 | |
| MYB | (35) | 185/198 | 185/212 | 0.93 | 0.87 | |
| NAC | (36) | 100/100 | 100/104 | 1.00 | 0.96 | |
| SBP | (37) | 16/17 | 16/16 | 0.94 | 1.00 | |
| WRKY | (38) | 71/72 | 71/72 | 0.99 | 0.99 | |
| OSAJ | bHLH | (39) | 134/166 | 134/143 | 0.81 | 0.94 |
| bZIP | (28) | 82/92 | 82/90 | 0.89 | 0.91 | |
| C2C2-GATA | (30) | 18/19 | 18/27 | 0.95 | 0.67 | |
| C3H | (31) | 65/67 | 65/70 | 0.97 | 0.93 | |
| MYB | (35) | 145/156 | 145/196 | 0.93 | 0.74 | |
| SBP | (37) | 18/19 | 18/19 | 0.95 | 0.95 |
The sensitivity and the PPV were determined for selected A. thaliana (ATH) and O. sativa ssp. japonica (OSAJ) TF families. For the PPV, a deviation from 1.00 means the inclusion of FPs. For the sensitivity, deviations from 1.00 indicate exclusion of true members (FNs). Families with both values larger than 0.90 appear in bold face. TPs according to gold standard.
MAIN RESULTS
In the current version of PlnTFDB (v3.0), we present a total of 84 different TF and TR families that occur in 19 different plant species and encompass 26 184 distinct proteins. A summary of the content of the database is shown in Table 1; there is a tendency that the number of TFs and TRs per family, as well as the number of families, increases along with the organismic complexity. Correlation analyses support this observation (Supplementary Data, Appendix 3).
The wide spectrum of gene families covered by PlnTFDB has already been exploited by researchers, e.g. for use in genome annotations (12,40,41), functional studies of TFs and TRs (42,43) and detailed phylogenetic studies of TF families in the whole plant lineage (28), among others.
OUTLOOK
As the cost of genome sequencing continues to decrease, the number of newly sequenced genomes will increase dramatically in the near future. The computational analysis pipeline behind PlnTFDB will be applied to these new genomes, increasing even further its wide phylogenetic coverage. We envisage that PlnTFDB will increasingly be exploited in genome annotation projects as a primary repository serving the identification of transcription regulatory proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Bundesministerium fuer Bildung und Forschung, Germany (GABI-FUTURE grant 0315046, GoFORSYS grant 0313924 and FRISYS grant 0313921); Subdirección de Investigación: Línea 15, Colegio de Postgraduados, México; Deutscher Akademischer Austauschdienst (DAAD). Funding for open access charge: GoFORSYS.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to express our gratitude to the people and institutions working on the sequencing and annotation of the plant genomes analyzed in this study. We are particularly thankful to Andreas Weber and Detlef Weigel who allowed us to explore plant genome data not published yet.
REFERENCES
- 1.Kummerfeld SK, Teichmann SA. DBD: a transcription factor prediction database. Nucleic Acids Res. 2006;34:D74–D81. doi: 10.1093/nar/gkj131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo AY, Chen X, Gao G, Zhang H, Zhu QH, Liu XC, Zhong YF, Gu X, He K, Luo J. PlantTFDB: a comprehensive plant transcription factor database. Nucleic Acids Res. 2008;36:D966–D969. doi: 10.1093/nar/gkm841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E. AGRIS and AtRegNet. A platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 2006;140:818–829. doi: 10.1104/pp.105.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riaño-Pachón DM, Ruzicic S, Dreyer I, Mueller-Roeber B. PlnTFDB: an integrative plant transcription factor database. BMC Bioinformatics. 2007;8:42. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yilmaz A, Nishiyama MY, Jr, Fuentes BG, Souza GM, Janies D, Gray J, Grotewold E. GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol. 2009;149:171–180. doi: 10.1104/pp.108.128579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richardt S, Lang D, Reski R, Frank W, Rensing SA. PlanTAPDB, a phylogeny-based resource of plant transcription-associated proteins. Plant Physiol. 2007;143:1452–1466. doi: 10.1104/pp.107.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmer A, Lang D, Richardt S, Frank W, Reski R, Rensing SA. Dating the early evolution of plants: detection and molecular clock analyses of orthologs. Mol. Genet. Genomics. 2007;278:393–402. doi: 10.1007/s00438-007-0257-6. [DOI] [PubMed] [Google Scholar]
- 8.Matsuzaki M, Misumi O, Shin IT, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 9.Barbier G, Oesterhelt C, Larson MD, Halgren RG, Wilkerson C, Garavito RM, Benning C, Weber AP. Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol. 2005;137:460–474. doi: 10.1104/pp.104.051169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worden AZ, Lee JH, Mock T, Rouze P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 11.Palenik B, Grimwood J, Aerts A, Rouze P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl Acad. Sci. USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 14.Zhao W, Wang J, He X, Huang X, Jiao Y, Dai M, Wei S, Fu J, Chen Y, Ren X, et al. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32:D377–D382. doi: 10.1093/nar/gkh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, Hamilton J, Haas B, Sultana R, Cheung F, et al. The institute for genomic research Osa1 rice genome annotation database. Plant Physiol. 2005;138:18–26. doi: 10.1104/pp.104.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 17.Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. The genome of black cottonwood, Populus trichocarpa (Torr& Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 20.The French Italian Public Consortium for Grapevine Genome Characterization. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 21.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duncan L, Nishii I, Harryman A, Buckley S, Howard A, Friedman NR, Miller SM. The VARL gene family and the evolutionary origins of the master cell-type regulatory gene, regA, in Volvox carteri. J. Mol. Evol. 2007;65:1–11. doi: 10.1007/s00239-006-0225-5. [DOI] [PubMed] [Google Scholar]
- 23.Dolinski K, Botstein D. Orthology and functional conservation in eukaryotes. Annu. Rev. Genet. 2007;41:465–507. doi: 10.1146/annurev.genet.40.110405.090439. [DOI] [PubMed] [Google Scholar]
- 24.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 25.Feng JX, Liu D, Pan Y, Gong W, Ma LG, Luo JC, Deng XW, Zhu YX. An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 2005;59:853–868. doi: 10.1007/s11103-005-1511-0. [DOI] [PubMed] [Google Scholar]
- 26.Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 2004;135:1738–1752. doi: 10.1104/pp.104.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Werber M, Weisshaar B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell. 2003;15:2497–2502. doi: 10.1105/tpc.151140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corrêa LG, Riaño-Pachón DM, Schrago CG, dos Santos RV, Mueller-Roeber B, Vincentz M. The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS ONE. 2008;3:e2944. doi: 10.1371/journal.pone.0002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003;3:17. doi: 10.1186/1471-2148-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyes JC, Muro-Pastor MI, Florencio FJ. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004;134:1718–1732. doi: 10.1104/pp.103.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Guo Y, Wu C, Yang G, Li Y, Zheng C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics. 2008;9:44. doi: 10.1186/1471-2164-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218:683–692. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Castilla LP, Alvarez-Buylla ER. Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc. Natl Acad. Sci. USA. 2003;100:13407–13412. doi: 10.1073/pnas.1835864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- 36.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 37.Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008;418:1–8. doi: 10.1016/j.gene.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 38.Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riaño-Pachón DM, Corrêa LG, Trejos-Espinosa R, Mueller-Roeber B. Green transcription factors: a Chlamydomonas overview. Genetics. 2008;179:31–39. doi: 10.1534/genetics.107.086090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, Fitzgerald LM, Vezzulli S, Reid J, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S. A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods. 2007;3:7. doi: 10.1186/1746-4811-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Street NR, Sjodin A, Bylesjo M, Gustafsson P, Trygg J, Jansson S. A cross-species transcriptomics approach to identify genes involved in leaf development. BMC Genomics. 2008;9:589. doi: 10.1186/1471-2164-9-589. [DOI] [PMC free article] [PubMed] [Google Scholar]