Abstract
IntAct is an open-source, open data molecular interaction database and toolkit. Data is abstracted from the literature or from direct data depositions by expert curators following a deep annotation model providing a high level of detail. As of September 2009, IntAct contains over 200.000 curated binary interaction evidences. In response to the growing data volume and user requests, IntAct now provides a two-tiered view of the interaction data. The search interface allows the user to iteratively develop complex queries, exploiting the detailed annotation with hierarchical controlled vocabularies. Results are provided at any stage in a simplified, tabular view. Specialized views then allows ‘zooming in’ on the full annotation of interactions, interactors and their properties. IntAct source code and data are freely available at http://www.ebi.ac.uk/intact.
INTRODUCTION
Proteins rarely fulfill their function in isolation, they typically form transient or stable interactions with other molecules. Determination and analysis of such interactions are essential to our biological and biomedical understanding of cellular function. Reflecting the importance of such information, a large number of experimental approaches are used to determine molecular interactions, yielding from very few to tens of thousands of interactions per published experiment. Experimental approaches can differ in the type of interaction data they generate. Some technologies, such as yeast two hybrid (1), generate pairs of interactors or binary interactions. Others, for example X-ray crystallography or tandem affinity purification (2), typically provide information on associations of more than two proteins. As with almost any scientific observation, the different methods do not necessarily report the biological ‘truth’, but may generate errors, of which false positives (reported interactions not relevant in vivo) and false negatives (in vivo relevant interactions not observed) are the most discussed in interactomics.
IntAct CURATION
For the correct interpretation of an interaction report in a database, it is desirable not only to have access to a high level of detail concerning the experimental technologies and conditions, but also to detailed biological results such as a description of the protein domains required to establish the interaction. Thus, IntAct follows a deep curation model, capturing a high level of detail from the experimental reports on the full text of the publication. We aim to provide the user with all the relevant experimental detail described in the originating article, with all entries being fully compliant with the International Molecular Exchange consortium (IMEx) (3) guidelines and the Minimum Information required to report a Molecular Interaction Experiment (MIMIx) standard (4), whilst also providing extra levels of information beyond these minimum requirements. To do so, IntAct makes extensive use of a number of controlled vocabularies, primarily the Molecular Interaction ontology of the Proteomics Standard Initiative (PSI-MI) (5) to describe the technical details of the experiment, binding sites, protein tags and mutations. The Gene Ontology (6) is used to describe the sub-cellular location an interaction may be shown to occur in or the function of an enzyme in an enzyme/substrate assay. Interacting molecules are systematically mapped to stable identifiers from public databases such as UniProtKB for proteins (7), ChEBI for small molecules (8), Ensembl for genes (9) and the DDBJ (10)/EMBL (11)/GenBank (12) nucleotide databases for nucleic acids. Features within a molecule, such as a binding site on a protein, are mapped to the sequence/structure given in the underlying database and remapped should a new version of the underlying sequence be released. Binding sites are also cross-referenced to the InterPro database (13), whenever possible. The full IntAct curation approach is documented in the publicly available IntAct curation manual (http://www.ebi.ac.uk/∼intact/site/doc/IntActAnnotationRules.pdf).
Database content
As with the data model of the PSI-MI standard, the basic curation unit in IntAct is the experiment, which is defined by a set of interactions reported in the same publication and derived using the same experimental technology. One publication might result in several experiment records, for example, if the authors initially used a high throughput technology to identify interactors and then confirmed selected interactions with a different technology. Throughout this document, we will use the term interaction for a set of two or more interacting molecules, and interaction evidence for a report of this interaction based on one technology in one publication.
The IntAct data model and XML downloads store n-ary interactions, which may appertain to a single interactor (in the case of a self-interaction) to many interactors in the case of large complexes. However, for many purposes, e.g. graphical representation of interaction networks, or comparison of data from multiple sources, it is necessary to generate binary interactions from complexes with more than two participants. IntAct does this following the spoke model (Supplementary Figure S1). As an example, an interaction of four interactors would result in three binary interaction evidences. As of September 2009, IntAct contains 200 056 binary interaction evidences abstracted from 4035 scientific publication. The increase of the number of evidences since the inception of the database can be observed in Supplementary Figure S2.
Interactions from over 275 species are described, although our main focus is on model organisms such as Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Escherichia coli and Arabidosis thaliana. Datasets for all these organisms can be retrieved from the IntAct FTP site or searched by common name or TaxID on the website.
Targeted datasets
IntAct continues to provide datasets on specific curation topics, either performing targeted curation for collaborators, such as on nucleoside diphosphate kinases (14), cyanobacteria (15) or Alzheimer’s Disease (Perreau et al., in preparation) or supporting areas of general interest, such as cancer (Derow et al., in preparation), epigenetics and apoptosis. Again, these datasets may be specifically searched or downloaded from the website. We are always interested in collaborations concerning targeted curation, if you are interested in working with us please contact the IntAct help desk.
Curated complexes
Sets of manually curated, stable protein complexes are being made available as a dictionary of complex nomenclature, with protein content clearly defined and linked to experimental evidence, with each variation of a complex given a distinct name and a separate entry. The function of each complex is given, synonyms collected, physical properties (if known) described and each complex has full Gene Ontology annotation, with additional links to the IntEnz (16), wwPDB (17) and Reactome databases (18), if applicable.
Direct data deposition
Most IntAct data content is derived from curation of the published literature. However, an increasing percentage of the data is derived from direct data depositions from authors as an integral part of the publication process, 8953 binary interaction evidences were submitted from January 2008 to September 2009. While an accession number will be issued immediately for submitted data, the data itself will be kept confidential until acceptance or publication of the manuscript. For details on data deposition in IntAct or one of its IMEx partner databases, please see http://www.ebi.ac.uk/intact/pages/documentation/data_submission.xhtml.
NEW IntAct WEBSITE
As the data accumulated in the IntAct database continues to grow and user requirements become more complex, a new website has been developed to meet the demands of an increased content load and to provide richer functionalities for searching, browsing and visualizing molecular interaction data. Some fundamental changes have been made in the way we present data, central to which is the initial presentation of the data as a list of binary interaction evidences. Users may now access the individual evidences that describe the interaction of two specific molecules, thus allowing users to filter result sets (e.g. by interaction detection method) to only retain user-defined evidences. For convenience purposes evidence pertaining to the same interactors are grouped together in the binary interaction evidence table.
In order to improve navigability, we have reorganized the layout of the website and grouped functionality in tabs enabling rapid access to specific sections of the web site. The following sections reflect the tabs available.
Home page
The home page provides general documentation, a simple search facility and a featured dataset of the month. The dataset of the month is usually emphasising a recent publication with directly submitted data, or a group of related publications curated in our targeted curation projects. The side bar menu gives easy access to both data and software downloads, documentation and statistics.
Search
In addition to a simple Google-like text search facility, the IntAct website allows the use of the Molecular Interaction Query Language (MIQL) (Aranda et al., in preparation) to search data. This provides a set of predefined fields that can be combined, enabling the creation of complex queries via use of boolean logic. Users can use and combine these fields manually in the search field. Additionally, one can get assistance by opening the Advanced Field search and accessing a list of existing fields and, where appropriate, a list of existing values (e.g. datasets, expansion methods). Upon adding fields to the current query, the list of resulting interactions gets updated.
More specialized searches can be accessed from the search tab. Users can now search for interactors that are annotated with terms from the Gene Ontology (19), InterPro, ChEBI and the UniProt Taxonomy, or interactions described using specific terms from the PSI-MI controlled vocabularies. Relevant suggestions are displayed while the user is typing a term name. Selecting a term in the suggestion list will display the matching set of interactions in the corresponding tab.
As IntAct now contains a growing set of interaction involving chemical compounds, a dedicated search module allowing users to search by chemical structure has been developed in collaboration with the ChEBI team. By drawing a chemical structure in the provided applet (requires a Java enabled browser), one can search for matching (exact, by similarity or substructure) molecules involved in IntAct interactions. Once the drawn structure is submitted, a matching list of molecules is displayed, each of which can then be selected to display interactions involving this compound.
Interactions
Once a query has been submitted to the website, the ‘Interactions’ tab automatically opens and presents the list of interactions matching the query (Figure 1). This table can be customized by clicking on the button ‘Change columns displayed’ in the table header so that one can choose the columns to be shown. When a large number of interactions match the query, the table will only show 30 interactions at a time and the user can browse subsequent pages using the navigation items in the table header. If the queried dataset contains spoke expanded interactions, a message will inform the user (above the table) and give the opportunity to easily filter these out. Should users wish to download a given dataset, it is possible to select one of the standard format such as PSI-MI XML or MITAB from the table header’s drop down menu and click on the export button to retrieve a data file.
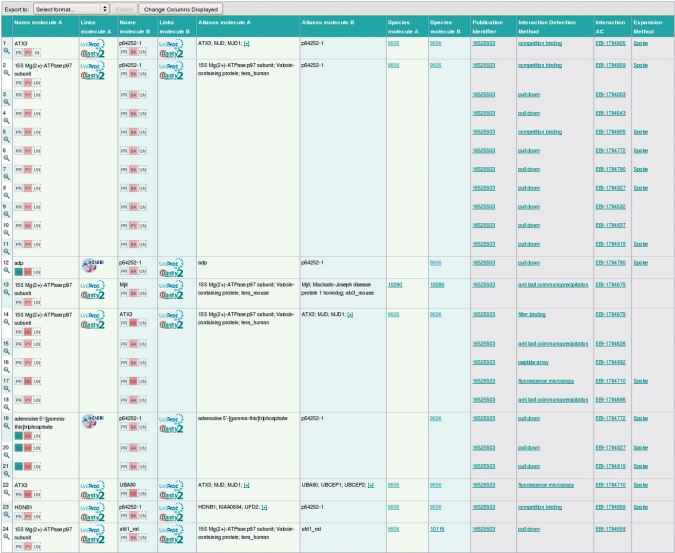
Figure 1.
The figure shows the interactions found after searching for Pubmed 16525503. Each row corresponds to one binary interaction with a list of evidences. Only the evidences matching the search criteria are shown in the results. In September 2009, 24 binary interaction evidences were found, 14 of which were originated from complexes and expanded using the Spoke model. Icons in the Name molecules fields show the type of molecule and the experimental and biological roles of the molecule in the context of the interaction. Links to Dasty2 and external resources such as UniProtKB or ChEBI are available too. Clicking on the magnifying glass icon in the first column or on the interaction accession number will direct the user to the details of that specific interaction evidence.
Clicking the magnifying glass in the far left-hand column will show to the user the detailed content of each interaction, which is explained in the section Interaction Details below.
Each participating molecule contains its ‘Links’ column, that contains the clickable logos to navigate to the main external resource about this molecule (e.g. UniProtKB or ChEBI). For proteins, a Dasty2 (20) logo can be clicked to navigate to the ‘Molecule View’ tab, explained below.
Browse
One of the new features of the website is the possibility of browsing result sets by selected topics. The different options are classified in groups, according to the participant molecule type: Proteins, Chemicals, or Nucleic Acids.
Based on the UniProt Taxonomy or Gene Ontology annotation of protein interactors, the current result set can be analysed/refined using a hierarchical display of ontology terms and the numbers of matching interactions in the current result set (Supplementary Figure S3). Other options link to external sites, and allow the visualization of InterPro domains, the involvement of molecules in Reactome pathways, the chromosomal location of the originating gene in Ensembl, or links to mRNA expression with ArrayExpress (21). To avoid overloading external sites, the number of molecules that IntAct uses in the links has been globally reduced to 200. When this happens, a corresponding warning message is shown so the user can either choose to narrow the result set or browse through the first 200 results.
For chemicals, the ChEBI ontology browser is available, which classifies the interactors according to Chemical Ontology terms such as role, molecular structure or subatomic particle.
Lists
The aim of the ‘Lists’ tab is to display the list of participants involved in the interactions from the result set. The molecules are classified by type. For each type, a table is shown listing the participants with selected attributes, such as the originating species or a description (Supplementary Figure S4). Depending on the molecule type, different actions can be performed using a custom selection of participants from the table. Most of the available options link to external sites using cross references as in the ‘Browse’ tab. This view also allows to expand the current result set, selecting some or all of the current interactors, and then searching all interactions these interactors participate in. From this view, it is possible to select specific molecules and search for the interactions in which they participate.
Interaction details
The interaction detail tab only becomes active when a single interaction has been selected from the interaction tab. This view shows the details of a given interaction and features additional manually annotated information, such as the binding domains, tags, and/or stoichiometry of the participants (Supplementary Figure S5).
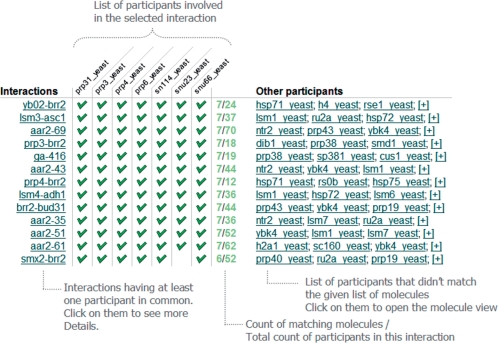
From here it is possible to find ‘similar’ interactions (i.e. interactions in which one or more of the participants of the interaction of interest have been also reported to interact). This may, for example, give additional evidence of the existence of a complex, suggest the architecture of a complex at the binary level, or give further interactions made by either a complex as a whole or by its components. The similar interactions are shown in a matrix where the columns are the participants and the rows are the interactions (Figure 2).
Figure 2.
The figure explains the ‘Find similar interactions’ matrix, which is accessible from the Details page.
Molecule view
In the tables from the ‘Interactions’ and ‘Lists’ views, clicking on the Dasty2 logo that can be found adjacent to protein molecules will redirect the user to the ‘Molecule View’ tab (see Supplementary Figure S6). Using the DAS protocol (22), this view shows protein features projected onto its sequence and, where available, its structure. It combines external data from UniProtKB, PDB, InterPro, among others, with IntAct data on e.g. interacting domains. Thus it is possible to obtain a visualization of, for example, an IntAct-annotated binding domain, its overlap with an InterPro domain and the localization of both features on a PDB structure, although none of the involved databases alone contains all the relevant data.
Graph—network visualization
One of the most popular features of interaction databases is the rendition of a graphical network derived from a selected set of interactions. IntAct provides a simple static graph under the ‘Graph’ tab, but interoperability with external visualization tools, such as the open source tool Cytoscape (23), has now become a priority. This has been made easily possible by the use of the PSI-MI standards by both IntAct and Cytoscape. Cytoscape can be started directly from within the IntAct website by clicking on the Cytoscape icon in the ‘Graph’ tab. The first time this is done it may take some time while the application is automatically downloaded and started. The ‘Graph’ tab is only active when the result set is relatively small (the current limit is set at 300 binary interactions) to avoid the complications related to data visualization of big networks. On the left-hand side, this view shows a simple representation of the network, where each line represents an interaction between two molecules. On the right of the page, the link to start Cytoscape on the fly using Java WebStart can be found.
Larger networks can be visualized by using a local installation of Cytoscape and exporting the interactions of interest using the Export but the ‘Interactions’ tab.
IMPLEMENTATION AND LOCAL INSTALLATION SUPPORT
IntAct is implemented using the Java language using a number of external and internal open source libraries. All the software produced by the IntAct developers is free and Open Source, and can be used, modified and redistributed under the terms of the Apache Software License. To further support reuse of IntAct software, we avoid dependencies involving external commercial libraries and tools wherever possible.
The IntAct model is defined by a database schema that is generic to enable its use by third parties. One such case is the MINT database (24), which uses the same model and libraries as part of its core implementation, but displays the data in a different way. The model and APIs are designed to be flexible, in order to all or a subset of the libraries to be used in other projects. To encourage its use, and build a user-community around the API, a public mailing list has been created to support its users and discuss development issues (http://groups.google.com/group/intact-developers).
In order to help new developers adopt and use the IntAct API, a project called intact-kickstart exists. This project contains examples demonstrating how to perform the most common tasks, for instance, how to insert data into a local database, how to work with files in the PSI-MI formats, or how to access IntAct data programmatically. This kickstart project is updated as new features are created and can be downloaded following the instructions at http://code.google.com/p/intact/wiki/GettingStarted. A guide on how to perform a local installation can be found in the IntAct developer pages (http://code.google.com/p/intact/wiki/LocalInstall).
For developers who do not use Java as their programming language, the support is limited to a number of libraries that can use PSI-MI XML as an input format, for example, the standalone RIntact R package (25), Bioconductor’s RpsiXML package (26) or the bioperl-network package for Perl (27). Another option is remote access to data from IntAct and a number of other interaction data resources using an implementation of the distributed PSICQUIC system (Aranda et al., in preparation). As it is a web service, any language can be used for this task.
FUTURE DEVELOPMENT
MITAB 2.6 format
Following the extensive feedback received for the first implementation of MITAB25 a new version of the format has been defined and specified. Basically, the new version is an extension of the current one, adding more standard columns to the data file with additional information about the binary interactions. For instance, columns containing the experimental methods, roles of the molecules or type of binary expansion (when the interaction is part of a complex) will be added.
New curation tool
Currently, IntAct is using a curation tool developed more than five years ago and a new tool is planned with features to match nowadays needs, hence increasing the productivity. As with the rest of IntAct web applications, this tool will be open source.
IMEx exchange
IntAct is already sharing the curation workload and avoiding redundant curation of interaction data in the public domain in the context of the IMEx consortium (http://imex.sf.net). We are currently implementing the next step in the IMEx collaboration, the regular exchange of curated data among IMEx partners.
Dynamic networks
Molecular interactions in living systems are clearly dynamic and dependent on factors such as sub-cellular localization or cell cycle phase. While we already annotate such factors where they are available from the experimental data, they are not yet accessible in a user-friendly way. We are currently conceptualising retrieval, browsing and analysis of dynamically changing interaction networks, both in textual and visual form.
DISCUSSION
In the domains of macromolecular structures and DNA sequences, deposition of experimentally derived data in public databases is a standard part of the scientific publication process. In interactomics, however, such practice is only slowly developing. As a result, many resources (28), including IntAct, aim to capture molecular interaction data from the scientific literature, through direct data deposition, expert curation, automated approaches like literature mining (29), or a combination of these (30); acting in isolation, or in coordination, for example as part of the IMEx database collaboration [DIP (31), IntAct (32), MINT, MPact (33), MatrixDB (34)]. Through IMEx we reduce redundant curation and increase overall coverage of published interaction data among public databases, and in collaboration with journal editors and the scientific community we aim to increase the proportion of interaction data that is directly deposited in public databases as part of the interaction process. However, to achieve the necessary change in common practice, we have to show the experimentalists who generate interaction data that first, molecular interaction databases are a valuable research tool which can increase the public visibility of their research results, and second, that the effort for data deposition in a public database is only marginal in comparison to the experimental effort originally required to generate the data. We hope that with the changes to the IntAct interface presented here, we have contributed to the first point, by increasing the value of IntAct as a research tool. For testing of the second point, we invite experimentalists generating interaction data to deposit their data in IntAct or another IMEx database as part of a journal submission, and to provide feedback on how we can further improve our data deposition and retrieval processes in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
IntAct is funded by the European Commission under FELICS, contract number 021902 (RII3) within the Research Infrastructure Action of the FP6 ‘Structuring the European Research’, under APO-SYS, contract number FP7-HEALTH-2007-200767, and under PSIMEx, contract number 223411 within the FP7-HEALTH-2007B programme. Funding for open access charge: EMBL-EBI.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Paula de Matos and the EBI ChEBI team for the development of the chemical search module in IntAct.
REFERENCES
- 1.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl Acad. Sci. USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 3.Orchard S, Kerrien S, Jones P, Ceol A, Chatr-Aryamontri A, Salwinski L, Nerothin J, Hermjakob H. Submit your interaction data the IMEx way: a step by step guide to trouble-free deposition. 2007;7(Suppl. 1):28–34. doi: 10.1002/pmic.200700286. [DOI] [PubMed] [Google Scholar]
- 4.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stümpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 5.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon—level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Gene Ontology Project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugawara H, Ikeo K, Fukuchi S, Gojobori T, Tateno Y. DDBJ dealing with mass data produced by the second generation sequencer. Nucleic Acids Res. 2009;37:D16–D18. doi: 10.1093/nar/gkn724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cochrane G, Akhtar R, Bonfield J, Bower L, Demiralp F, Faruque N, Gibson R, Hoad G, Hubbard T, Hunter C, et al. Petabyte-scale innovations at the European Nucleotide Archive. Nucleic Acids Res. 2009;37:D19–D25. doi: 10.1093/nar/gkn765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta A, Orchard S. Nucleoside diphosphate kinase (NDPK, NM23, AWD): recent regulatory advances in endocytosis, metastasis, psoriasis, insulin release, fetal erythroid lineage and heart failure; translational medicine exemplified. Mol. Cell Biochem. 2009;329:3–15. doi: 10.1007/s11010-009-0114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaut M, Kerrien S, Montecchi-Palazzi L, Chauvat F, Cassier-Chauvat C, Aude JC, Legrain P, Hermjakob H. InteroPORC: automated inference of highly conserved protein interaction networks. Bioinformatics. 2008;24:1625–1631. doi: 10.1093/bioinformatics/btn249. [DOI] [PubMed] [Google Scholar]
- 16.Fleischmann A, Darsow M, Degtyarenko K, Fleischmann W, Boyce S, Axelsen KB, Bairoch A, Schomburg D, Tipton KF, Apweiler R. IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 2004;32:D434–D437. doi: 10.1093/nar/gkh119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jimenez RC, Quinn AF, Garcia A, Labarga A, O'N;eill K, Martinez F, Salazar GA, Hermjakob H. Dasty2, an Ajax protein DAS client. Bioinformatics. 2008;24:2119–2121. doi: 10.1093/bioinformatics/btn387. [DOI] [PubMed] [Google Scholar]
- 21.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update—from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang T, Li N, Orchard S, Kerrien S, Hermjakob H, Gentleman R, Huber W. Rintact: enabling computational analysis of molecular interaction data from the IntAct repository. Bioinformatics. 2008;24:1100–1101. doi: 10.1093/bioinformatics/btm518. [DOI] [PubMed] [Google Scholar]
- 26.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JGR, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatr-aryamontri A, Kerrien S, Khadake J, Orchard S, Ceol A, Licata L, Castagnoli L, Costa S, Derow C, Huntley R, et al. MINT and IntAct contribute to the Second BioCreative challenge: serving the text-mining community with high quality molecular interaction data. Genome Biol. 2008;9(Suppl. 2):S5. doi: 10.1186/gb-2008-9-s2-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceol A, Chatr-Aryamontri A, Licata L, Cesareni G. Linking entries in protein interaction database to structured text: the FEBS Letters experiment. FEBS Lett. 2008;582:1171–1177. doi: 10.1016/j.febslet.2008.02.071. [DOI] [PubMed] [Google Scholar]
- 31.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Güldener U, Münsterkötter M, Oesterheld M, Pagel P, Ruepp A, Mewes H, Stümpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chautard E, Ballut L, Thierry-Mieg N, Ricard-Blum S. MatrixDB, a database focused on extracellular protein-protein and protein-carbohydrate interactions. Bioinformatics. 2009;25:690–691. doi: 10.1093/bioinformatics/btp025. [DOI] [PMC free article] [PubMed] [Google Scholar]