Abstract
Ensembl Genomes (http://www.ensemblgenomes.org) is a new portal offering integrated access to genome-scale data from non-vertebrate species of scientific interest, developed using the Ensembl genome annotation and visualisation platform. Ensembl Genomes consists of five sub-portals (for bacteria, protists, fungi, plants and invertebrate metazoa) designed to complement the availability of vertebrate genomes in Ensembl. Many of the databases supporting the portal have been built in close collaboration with the scientific community, which we consider as essential for maintaining the accuracy and usefulness of the resource. A common set of user interfaces (which include a graphical genome browser, FTP, BLAST search, a query optimised data warehouse, programmatic access, and a Perl API) is provided for all domains. Data types incorporated include annotation of (protein and non-protein coding) genes, cross references to external resources, and high throughput experimental data (e.g. data from large scale studies of gene expression and polymorphism visualised in their genomic context). Additionally, extensive comparative analysis has been performed, both within defined clades and across the wider taxonomy, and sequence alignments and gene trees resulting from this can be accessed through the site.
INTRODUCTION
Since 1995, when the genome of a cellular organism was completely sequenced for the first time, genome sequencing has transformed the biological sciences. Today, in excess of 1000 genomes have been sequenced, assembled, annotated and deposited in the public nucleotide archives; numerous other genomes exist in states of partial assembly and annotation; thousands of viral genomes sequences have also been generated. Moreover, the increasing use of high-throughput sequencing technologies is rapidly reducing the cost of genome sequencing, leading to an accelerating rate of data production. This not only makes it likely that in the near future, the genomes of all species of scientific interest will be sequenced; but also the genomes of many individuals, with the possibility of providing accurate and sophisticated annotation through the similarly low-cost application of functional assays. Many of these trends have first become visible in human genomics, but are spreading to other species as costs continue to fall. For example, the 1000 genomes project (to sequence 1000 human genomes) has quickly been followed by the launch of similar initiatives in Arabidopsis (1), Plasmodium (http://www.genome.gov/26523588), Drosophila (http://www.dpgp.org), and other species.
These developments place particular demands on biological databases, but also represent an opportunity. The European Bioinformatics Institute (EBI) is involved (together with our collaborators) in the operation of a number of archival databases, e.g. the European Nucleotide Archive (ENA) (2), ArrayExpress (3) (for gene expression data), PRIDE (4) (for proteomics data) etc. Such databases remain essential as a permanent record of experimental effort, and are under continuous development to accommodate data produced by new technologies. The UniProt Knowledgebase (UniProtKB) (5), in contrast, uses information curated from the scientific literature to describe proteins, which can subsequently serve as reference data for systems designed to automatically transfer annotation to similar, less well-characterised sequences. However, such computed annotation is best recalculated periodically as algorithms are refined and the quantity and quality of reference data increases. Moreover, the interpretation of potentially huge raw data sets as integrated features (e.g. a gene ‘built’ from transcriptional evidence) is a necessity if users are not to drown in an excess of information. This calls for a different type of resource i.e. derived and interpreted, complementing the archival and literature-based resources.
The structure of a genome provides a natural index through which data from other molecular biology experiments can be accessed. The Ensembl (6) platform for genome annotation and visualisation has been under development by the EBI and the Wellcome Trust Sanger Institute since 2000, with an initial focus on the human genome that has since widened to include other vertebrates. This paper describes ‘Ensembl Genomes’, a new resource recently launched by the EBI to complement Ensembl by providing access to genome-scale data from non-vertebrate species through the same user interfaces as Ensembl. There are two big advantages of this approach. First, the Ensembl software system has evolved in response to new data types that have frequently first appeared in human genomic studies, but which are now increasingly appearing in the context of other species; its re-use provides a cost-effective way of providing sophisticated data analysis and visualisation tools for a wider range of genomes. Secondly, the use of a common system for all genomes greatly decreases the cost of broad range comparative genomics and other inter-species data analysis. For example, since the launch of Ensembl Genomes, data from malarial parasites (genus Plasmodium, available through Ensembl Protists), the malarial vector (Anopheles gambiae, available through Ensembl Metazoa), and the human host (in Ensembl) can now be accessed through a common set of interfaces.
With Ensembl Genomes, we aim to provide an integrated portal to reference sequence and annotation from all species of scientific interest; and thereby to complement, and provide convenient access to, data from existing and novel community initiatives in particular areas. Many of the databases that support Ensembl Genomes have been built by, or in close collaboration with, groups who maintain specialist data resources for individual species, and we are actively seeking to extend the range of these collaborations. The development of accurate and useful genome databases is wholly dependent on the involvement of scientists with knowledge of the most important data and its biological context. Ensembl Genomes can support such expertise through the provision of a data analysis and service infrastructure, close integration with the archival databases, and a single unified interface across the taxonomic space.
Ensembl is not the only browser for genome data. For example, Gbrowse (7) can be run off a simple flat file, and is used by a variety of databases maintained by groups ranging from small research communities to genome sequencing centres; the UCSC Genome Browser Database (8) supports a similar range of invertebrate metazoan genomes to Ensembl Genomes; the NCBI (9) maintains a number of tools for genome browsing with comprehensive coverage across the taxonomic spectrum. Comparative strengths of the Ensembl platform include the variety of available interfaces and the relatively sophisticated representation of complex data (both in the database and in the interfaces), close integration with the international archive databases and, with the launch of Ensembl Genomes, breadth of taxonomic spread. Moreover, Ensembl is not just a data visualisation tool, but a suite of programs for data production (e.g. gene calling, comparative genomics) that can be deployed individually according to the needs of an individual community. We are looking forward to working with increasing numbers of collaborators to improve not only access to genome annotation, but the quality and depth of the annotation itself.
OVERVIEW OF ENSEMBL GENOMES
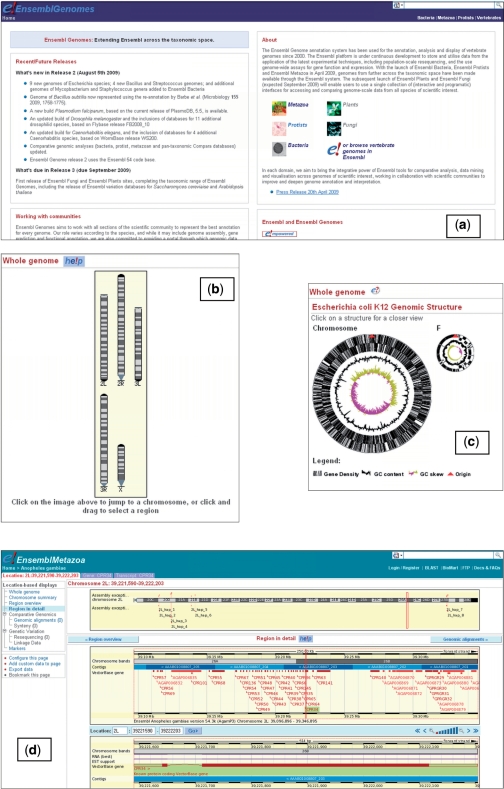
Ensembl Genomes (http://www.ensemblgenomes.org) consists of separate portals for each of five distinct domains of life: bacteria, protists, fungi, plants, and invertebrate metazoa. Each site contains data for selected species from within their domain, chosen for their scientific interest. Information is available about each species, including the assembly version and annotation methods used, the overall composition of the genome, etc. These have been customised for the species in Ensembl Genomes; for example, in Ensembl Bacteria, a circular view of the genome is provided, and the top-level pages have been designed to provide structured access to the large numbers of similar strains that have been sequenced in many clades. The main functionality offered by the portals is access to a graphical view of each genome, using the Ensembl genome browser software. The browser offers a number of views of locations in the genome, genes, and specific transcripts; tabs at the top of the page allow users to switch between these different levels, while a context-dependent left hand menu offers access to a selection of data views specific to each. The main display then appears in the central panel. Users can integrate their own analyses into appropriate displays by using the Distributed Annotated System (10) (DAS). Specific views exist for expression data, SNPs and other polymorphisms, and comparative genomics. With each new data release, the browser software is updated to a recent version, ensuring that a consistent user experience is provided between Ensembl and Ensembl Genomes. Release frequency is four to six times a year, tied to Ensembl releases where practical. Use of the browser is illustrated in Figure 1.
Figure 1.
Some views of Ensembl Genomes (a) the homepage (b) graphical karyotype view for Anopheles gambiae, using the same representation as found in vertebrate Ensembl (c) the equivalent view for a bacterial genome, showing an alternative representation more suitable for circular chromosomes and plasmids. This view has been produced using the program circular_diagram.pl, (ftp://ftp.sanger.ac.uk/pub/software/artemis/extra/circular_diagram.pl), which has been incorporated into the Ensembl browser (d) location view for a region of the left arm of chromosome 2 of A. gambiae, annotated by VectorBase. The image illustrates gene and (at a larger scale) transcript-based views. The views are track based, with features of particular types located in their own horizontal bands (which can be turned on or off using a control panel). On the lower panel, a single track (labelled ‘RNA best EST support’) provides the evidence for the transcript, which appears in a second track labelled ‘VectorBase gene’. A variety of alternative location-based views can be selected in the left hand column; the tabs (at the base of the banner) allow users to switch between location, gene and transcript-centric views.
In addition, Ensembl Genomes data is available through a number other routes. These include (i) query optimised data warehouses [built using the BioMart data warehousing system (11)] that can be accessed through a variety of interfaces including SOAP-based and RESTful web services (ii) a publicly accessible relational database server offering direct access to all Ensembl Genomes databases and which supports a Applications Programming Interface (API) for the Perl programming language (iii) an FTP site where data can be downloaded in bulk. DNA and protein sequence similarity search is provided using BLAST (12). The URLs by which these services can be accessed are shown in Table 1. These routes of access match those available for vertebrate data in Ensembl; all data is available for re-use without restriction. As Ensembl software is also freely distributed, users can also establish their own local mirrors, and view confidential data in the context of the public reference.
Table 1.
URLs for accessing Ensembl and Ensembl Genomes
| Ensembl (vertebrates) | http://www.ensembl.org |
|---|---|
| Ensembl Genomes | http://www.ensemblgenomes.org |
| Public mysql server | mysql -hmysql.ebi.ac.uk -P4157 -uanonymous |
| Ensembl Bacteria | |
| Web browser | http://bacteria.ensembl.org |
| FTP | ftp://ftp.ensemblgenomes.org/pub/bacteria/ |
| BioMart | http://bacteria.ensembl.org/biomart |
| Ensembl Protists | |
| Web browser | http://protists.ensembl.org |
| FTP | ftp://ftp.ensemblgenomes.org/pub/protists/ |
| BioMart | http://protsits.ensembl.org/biomart |
| Ensembl Fungi | |
| Web browser | http://fungi.ensembl.org |
| FTP | ftp://ftp.ensemblgenomes.org/pub/fungi/ |
| BioMart | http://fungi.ensembl.org/biomart |
| Ensembl Plants | |
| Web browser | http://plants.ensembl.org |
| FTP | ftp://ftp.ensemblgenomes.org/pub/plants/ |
| BioMart | http://plants.ensembl.org/biomart |
| Ensembl Metazoa | |
| Web browser | http://metazoa.ensembl.org |
| FTP | ftp://ftp.ensemblgenomes.org/pub/metazoa/ |
| BioMart | http://metazoa.ensembl.org/biomart |
All BioMarts are additionally available through BioMart central at http://www.biomart.org/biomart/martview/.
These various interfaces are supported by a number of relational (MySQL) database schemas; a core database (describing sequence, features and stable identifiers) and optional additional databases for certain types of satellite data. Generally, one database of each type is provided for each genome; however, comparative genomics databases can cover many species. Additionally, with the launch of Ensembl Bacteria, the Ensembl schema and API has been transparently extended to support the storage of multiple genomes in a single database schema; this is used to collect multiple (small) genomes of closely related species/strains into a single database, allowing the management of data for large numbers of genomes without an explosion in the total number of databases.
DATA IN ENSEMBL GENOMES
Ensembl Genomes has a varied relationship with the scientific community in the preparation of the databases that appear in the portal. In the case of some genomes, EBI staff have been active participants in their primary annotation. In other cases, we work actively with collaborators to build Ensembl databases. In other cases, we import canonical data from a generally-acknowledged authority for a given species and transform this into an Ensembl schema. For some species, there is no community authority actively curating annotations; in these cases, data may be imported from the records previously submitted to the ENA/GenBank/DDBJ public nucleotide archives by the sequencing consortium, supplemented by protein centric functional annotation imported from the UniProtKB.
Imported data typically includes DNA sequence and gene annotations; the quantity of depth of annotation varies with the species and source. Additional computed annotations (e.g. non-protein coding genes, repeat features, cross references, functional annotation inferred directly from sequence, and data from genome-wide functional assays) are selectively integrated with the imported data prior to release.
The specific content of the databases (as of release 2, August 2009) is described below.
Ensembl Bacteria
Ensembl Bacteria contains databases for six bacterial clades (Bacillus, Escherichia/Shigella, Mycobacterium, Neisseria, Streptococcus and Staphylococcus), and one archaeal clade (Pyrococcus), using records in the ENA/GenBank/DDBJ nucleotide sequence archives as the primary source of sequence and annotation. Each database contains many closely related genome sequences, ranging from four genomes in the Pyrococcus and Neisseria clades to 35 Streptococcus genomes. A total of 144 genome sequences are present in the database. The database structure is hidden from users of the genome browser, who are able to visualise each genome as if it were in a separate Ensembl database. Information about operons and co-transcribed units for Escherichia coli has been imported from Regulon DB (13).
Ensembl Protists
Ensembl Protists comprises databases for three malarial parasites. Data for Plasmodium falciparum, the primary species causing malaria in humans, has been imported from PlasmoDB (14), a database which integrates information from plasmodial genomes; databases from two other species (P. vivax and P. knowlesi) have been built from sequence and annotation in the ENA/GenBank/DDBJ nucleotide sequence archives.
Ensembl Fungi
Ensembl Fungi contains data for the budding yeast, Saccharomyces cerevisiae, the fission yeast Schizosaccharomyces pombe, and seven species of the genus Aspergillus. For S. cerevisiae, data has been imported from the Saccharomyces Genome Database (15) (SGD), based on SGD’s release of June 2009 and integrated with gene expression data from ArrayExpress and information from a recent survey of polymorphism (16). The S. pombe data is imported from GeneDB (17). The Aspergillus databases have been developed in partnership with the Central Aspergillus Data Repository (18), which already uses Ensembl technology for the management and visualisation of data.
Ensembl Plants
Ensembl Plants is being developed in collaboration with Gramene (19), a resource for plant comparative genomics already developed (at the Cold Spring Harbor Laboratory) using the Ensembl infrastructure. Ensembl Genomes and Gramene have commited to maintaining a common set of databases to be accessible via both sites; the initial release, is based on Gramene release 30. The initial release of Ensembl Plants includes the two genomes of four dicotylendons: Vitis vinifera, Populus trichocarpa, and two species of Arabidopsis; and four montocyledons:, Oryza sativa groups indica and japonica, Brachypodium distachyon, and Sorghum bicolor. Variation databases have been provided for three species: A. thaliana, O. sativa, and Vitis vinifera. The A. thaliana data is based on the TAIR9 assembly and is comprised of three data sets: the Perlegen 1 Million SNP set (20), the Affymetrix 250k SNP chip set (which is derived from Perlegen), and SNPs and indels derived from the resequencing of 17 accessions by Mott (University of Oxford) and Kover (University of Manchester). The grape data is based on the IGGP assembly and is comprised of a single data set, created from the re-sequencing of 11 V. vinifera cultivars and 6 wild Vitis species by Buckler and Ware (USDA ARS). The rice database is assembled using data imported from dbSNP (21). Gene and protein-based comparative analysis is also available.
Ensembl Metazoa
Ensembl Metazoa is built primarily from data maintained by three community-based genomics resources for different types of invertebrate metazoa: FlyBase (22) (focused on Drosophila); WormBase (23) (focused on Caenhorhaditis elegans and related nematodes) and VectorBase (24) (a NAIAD resource for invertebrate vectors of human pathogens). WormBase and VectorBase actively contribute to Ensembl Metazoa by building Ensembl databases themselves. The current release contains four vector genomes (Aedes aegytpii, Anopheles gambiae, Culex quinquefasciatus and Ixodes scapularis), five genomes of the Caenhorhabditis genus (including a new build of the C. elegans genome, based on WormBase release WS200), and 12 Drosophila genomes (including a new build of D. melanogaster) based on FlyBase release 2008_10). A variation database for A. gambiae is already available and a similar resource for D. melanogaster, based on data from the Drosophila Population Genome Project, is in development. Other species targeted for inclusion within the next 6 months include Pediculus humanus, Apis mellifera, Bombyx mori, Tribolium castaneum and Acyrthosiphon pisum.
COMPARATIVE GENOMICS
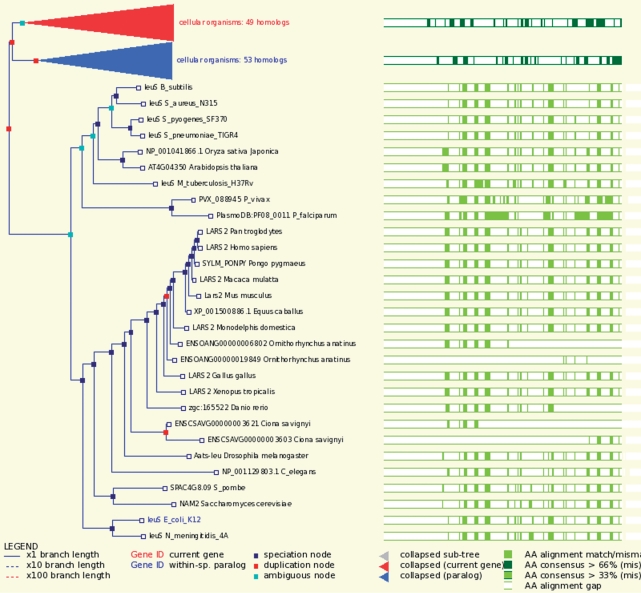
In each of the Ensembl Genomes sectors, comparative analysis is provided using the Ensembl Compara pipeline previously applied to vertebrate genomes and their outliers in Ensembl. Compara consists of two primary modules, one for DNA alignment (25) and one for protein alignment (26). The DNA alignment module uses a graph-based approach (Enredo) for identifying duplicated regions and specifying the input for Pecan, a high-performance multiple aligner; a third module, Ortheus, calculates ancestral sequences. The protein alignment module involves an all-against-all BLAST comparison, the derivation of clusters, and the interpretation of evolutionary trees for each cluster of related genes in the context of the known species tree, leading to the identification of gene duplication and speciation events. A typical gene tree is illustrated in Figure 2.
Figure 2.
The gene tree for valine, isoleucine and leucine synthetase, computed by the application of the Compara gene trees pipeline to 29 species from across the taxonomy. The branches for valine and isoleucine synthetase have been collapsed. An alignment is shown to the right of the tree.
The protein-based approach is applicable over a wide taxonomic scope and a pan-taxonomic database has also been produced, using a selection of species from across the Ensembl and Ensembl Genomes domains. The most recent analysis has been built using 357 332 genes from 29 species, including 12 vertebrates, 3 invertebrate metazoa, 2 plants, 2 fungi, 2 protists, 7 bacteria and 1 archaeon. A total of 26,081 clusters were generated, of which 25% (containing 62% of the genes) span 2 or more domains, 6% (14% of genes) span eukaryotic and non-eukaryotic species, and 1.6% (7% of genes) span all Ensembl domains. The average number of genes per cluster is 14 (from an average of 6 species), rising to 33 (from 14 species) for clusters spanning at least 2 domains, and 57 (from 24 species) from clusters spanning the entire taxonomy. New Compara databases are produced for each domain of Ensembl Genomes with each release, and the resulting alignments and trees can be visualised in the web browser and downloaded via FTP.
FUTURE PLANS
The Ensembl browser is a powerful tool for genome analysis and visualisation, but it was conceived with a relatively small number of vertebrate genomes in mind. In particular, work is underway to improve the interface specifically to address certain issues raised in the context of bacterial genomes, whose characteristics are in some ways markedly different to those of vertebrates. Features in development include improved visualisation for circular genomes and polycistronic transcripts (features of specific to bacterial genomes), and the introduction of improved interfaces for selection from large numbers of species when specifying the domain for search. We are also working on building the recognition of potential horizontal gene transfer events into the Compara pipeline for the interpretation of gene trees; and in representing the genomes of independently sequenced, but highly similar, isolates more efficiently and usefully as variants on a single reference sequence, instead of as independent reference genomes (the current default implementation).
Comparative genomics is an area where additional developments are planned. Together with Ensembl, we are working on changes to the Compara pipeline to improve the integration of broad and narrow scope comparative analysis, linking the pan-taxonomic Compara database to clade-specific databases, through the use of a common set of predictive models to identify the cluster members. We have also identified over 30 additional species (17 bacteria, 4 archaea, 6 protists, 2 fungi and 5 metazoan) as priorities for incorporation within Ensembl Genomes, for the purpose of improving the sampling of the major domains of life; and are working with other groups interested in comparative analysis to help establish a reference set of species for comparative analysis.
However, as the cost of sequencing a genome continues to fall, it is not realistic to expect that every species will be the focus of dedicated bioinformatic effort, simply because its genome has been sequenced. Instead, our main focus will be on improving the quality and depth of annotation in species of scientific interest, in partnership with communities who understand how genomics can help them address scientific problems, what data is most important to their work, and where there is most scope for (and value in) improvement in the existing annotation. We will continue to be interested on human pathogens and vectors: new areas of focus are likely to include plant pathogens, and important cereal crops whose genomes will start to be deciphered within the next two years.
FUNDING
European Molecular Biology Laboratory; the European Commission [under FELICS, 021902 (RII3)]; and the Biotechnology and Biosciences Research Council (BB/F019793/1). Funding for open access charge: The European Molecular Biology Laboratory (employer of primary author).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Kim Rutherford for permission to use circular_diagram.pl within EnsemblGenomes.
REFERENCES
- 1.Weigel D, Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 2009;10:107. doi: 10.1186/gb-2009-10-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cochrane G, Akhtar R, Bonfield J, Bower L, Demiralp F, Faruque N, Gibson R, Hoad G, Hubbard T, Hunter C, et al. Petabyte-scale innovations at the European Nucleotide Archive. Nucleic Acids Res. 2009;37:D19–D25. doi: 10.1093/nar/gkn765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update – from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones P, Côté RG, Cho SY, Klie S, Martens L, Quinn AF, Thorneycroft D, Hermjakob H. PRIDE: new developments and new datasets. Nucleic Acids Res. 2008;36:D878–D883. doi: 10.1093/nar/gkm1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenkinson A, Albrecht M, Birney E, Blankenburg H, Down T, Finn R, Hermjakob H, Hubbard T, Jimenez R, Jones P, et al. Integrating biological data – the Distributed Annotation System. BMC Bioinformatics. 2008;9(Suppl. 8):S3. doi: 10.1186/1471-2105-9-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart – biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Gama-Castro S, Jiménez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Peñaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muñiz-Rascado L, Martínez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mabey Gilsenan JE, Atherton G, Bartholomew J, Giles PF, Attwood TK, Denning DW, Bowyer P. Aspergillus genomes and the Aspergillus cloud. Nucleic Acids Res. 2009;37:D509–D514. doi: 10.1093/nar/gkn876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T, Hurwitz B, McCouch S, Ni J, Pujar A, et al. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 2008;36:D947–D953. doi: 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. Common Sequence Polymorphisms Shaping Genetic Diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 21.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drysdale R. FlyBase: a database for the Drosophila research community. Methods Mol. Biol. 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. [DOI] [PubMed] [Google Scholar]
- 23.Bieri T, Blasiar D, Ozersky P, Antoshechkin I, Bastiani C, Canaran P, Chan J, Chen N, Chen WJ, Davis P, et al. WormBase: new content and better access. Nucleic Acids Res. 2007;35:D506–D510. doi: 10.1093/nar/gkl818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:D583–D587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18:1814–1828. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]