Abstract
Ensembl (http://www.ensembl.org) integrates genomic information for a comprehensive set of chordate genomes with a particular focus on resources for human, mouse, rat, zebrafish and other high-value sequenced genomes. We provide complete gene annotations for all supported species in addition to specific resources that target genome variation, function and evolution. Ensembl data is accessible in a variety of formats including via our genome browser, API and BioMart. This year marks the tenth anniversary of Ensembl and in that time the project has grown with advances in genome technology. As of release 56 (September 2009), Ensembl supports 51 species including marmoset, pig, zebra finch, lizard, gorilla and wallaby, which were added in the past year. Major additions and improvements to Ensembl since our previous report include the incorporation of the human GRCh37 assembly, enhanced visualisation and data-mining options for the Ensembl regulatory features and continued development of our software infrastructure.
INTRODUCTION
On 27 January 2000, the Ensembl project announced the completion of ‘Ensembl Milestone 1’, the first complete release of the project’s data and web interface. The release included gene predictions and repeat annotations across all human DNA sequence available at that time (both finished and draft) and provided supporting evidence for the gene predictions including protein homology matches. All data was accessible though the Ensembl web site and via FTP download. In the email that announced Milestone 1, we noted that work on Ensembl was still in progress and promised significant improvements in the coming months.
Ten years later, Ensembl provides a much larger and more complete genomic information resource in support of dozens of genomes including gene sets, multi-species alignments, annotations of orthologous and paralogous genes, and extensive variation and regulatory information. All data is available through a variety of visual and programmatic interfaces including the Ensembl Genome Browser, the Perl API and BioMart. We also provide a complete copy of all data and code to be used freely by the community.
Ensembl works closely with a number of other fundamental bioinformatics projects that provide resources to the wider research community to ensure data consistency and increase resource interconnectedness. Some of these projects include the Genome Browser at UCSC (1), the databases and resources of the NCBI (2), the Rat Genome Database (RGD) (3) and VEGA (4).
In this article, we provide a general overview of the data available within Ensembl and highlight some of the features and developments that have been introduced since our last report (5). Ensembl is comprehensively updated approximately five times each year and details of the new and updated data in each release are always provided on the Ensembl news pages linked from http://www.ensembl.org. Additionally, we provide more immediate information on the Ensembl blog at http://ensembl.blogspot.com/ as well as through the low volume Ensembl announce mailing list, which is open to all. To subscribe to the list, send an email to majordomo@ebi.ac.uk with the text ‘subscribe ensembl-announce’ as the message body.
RESULTS
Over the past year, we introduced seven new species including the anole lizard (Anolis carolinensis), the first reptile in Ensembl. Other species included the two-toed sloth (Choloepus hoffmanni), white-tufted-ear marmoset (Callithrix jacchus), the pig (Sus scrofa), the Tammar wallaby (Macropus eugenii), the zebra finch (Taeniopygia guttata) and the Western lowland gorilla (Gorilla gorilla). Of these, the anole lizard, zebra finch, marmoset and pig were high coverage genome assemblies based on ∼4–6 times coverage from Sanger-style sequencing reads and gorilla was the first example of an assembly that combined traditional Sanger-style sequencing at low coverage with high-throughput short-read sequencing at high coverage. Ensembl now fully supports a total of 24 high-coverage chordate genomes and 23 low-coverage chordate genomes. The lamprey (Petromyzon marinus), another high-coverage chordate genome, is currently provided with preliminary support only. An additional three non-chordate species (Saccharomyces cerevisiae, Caenorhabditis elegans and Drosophila melanogaster) are included to facilitate comparative analysis.
As of release 56 (September 2009), we transferred support of two mosquito species from Ensembl to our sister project Ensembl Genomes (http://www.ensemblgenomes.org). Both aedes (Aedes aegypti) and anopheles (Anopheles gambiae) will continue to be available through Ensembl Metazoa (http://metazoa.ensembl.org).
Gene annotation
In addition to the newly supported species listed above, for each of which we released a comprehensive gene set, we released new gene sets for a number of other species. In general new gene sets are released in conjunction with each new genome assembly. Our largest single effort over the past year has been in support of the GRCh37 human assembly, which was released by the Genome Reference Consortium in early 2009 (http://www.genomereference.org). This new build includes a long list of genomic regions that have been assessed for accuracy and updated where necessary. To support projects such as ENCODE and the 1000 Genomes, we will continue to provide complete resources for the NCBI36 human assembly in the form of an enhanced Ensembl archive site including BLAT/BLAST sequence search and other features not present in standard archive sites. The site, http://ncbi36.ensembl.org, will remain active until at least Summer 2010 when, depending on usage, we intend to provide support for the NCBI36 assembly only in the form of a typical Ensembl archive site.
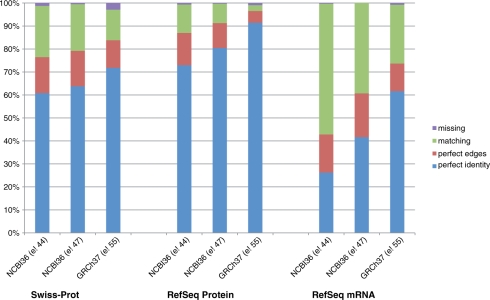
With the new GRCh37 assembly, a larger fraction of Ensembl genes correspond to RefSeq (6) and UniProt (7) entries suggesting continuing convergence of all of these resources [Figure 1 and compare with Figure 1 in Birney et al. (8)]. The improved convergence level is the result of at least three components: First, the genome assembly has improved. Second, the Ensembl gene build strategy has improved including the development of a combined Ensembl/Havana merged gene set (5), which increased the number of protein-coding transcripts. Third, the other resources (i.e. RefSeq and UniProtKB) have themselves independently improved their quality and internal consistency.
Figure 1.
The convergence of the Ensembl gene set and the UniProt and RefSeq resources shown over time. Three versions of Ensembl (release 44 in April 2007, release 47 in October 2007 and release 55 in July 2009) are each compared to the data available from Swiss-Prot/UniProtKB, NCBI RefSeq Proteins and NCBI RefSeq mRNAs. The colours on the bars represent the fraction of the Ensembl entries that perfectly match the entries in the other resources (blue); have matching edges and an internal mismatch or indel (red); have a substantial, but incomplete match (green); or are missing (purple).
Ensembl, in partnership with NCBI, UCSC and the Havana project, continues to play an active role in the CCDS consortium (9). As of Ensembl release 56 (September 2009), 19 851 Ensembl translations match human CCDS consensus coding region structures exactly, and 17 679 Ensembl translations match mouse CCDS structures exactly. In last year’s report, we described in detail the creation of the extensively supported human and mouse genes sets through the merging of the Ensembl and Havana gene sets. These efforts continue in the context of the GENCODE project and have culminated in the Ensembl release 56 geneset becoming the GENCODE geneset (release 3c).
Beyond human and mouse, we released a new gene set in support of the Zv8 assembly of the zebrafish genome, which incorporates many of the new methods applied in the human and mouse builds. For the rat genome, we released a completely updated gene set using the previous assembly to incorporate the significant additional supporting information that had become available since our previous gene set was created. A number of other species, including horse and cow, received relatively minor updates. Ensembl also incorporated the data formerly held in the Alternative Splicing and Transcript Diversity (ASTD) database as part of the planned decommissioning of this database and consolidation of genomic annotation data (10).
Functional genomics and regulatory information
We have continued development of the Ensembl Regulatory Build that has been briefly described in our previous reports (5,11). Over the past year we released two updates to the set of human Ensembl regulatory features and continued our focus on CD4+T-cells by incorporating additional histone modification data from Wang et al. (12). We also released the first version of the mouse regulatory build focused on embryonic stem (ES) cells based in part on data from Mikkelson et al. (13). Additionally, Ensembl regulatory features and their supporting data such as sites of DNase I hypersensitivity and selected histone modifications are now available via BioMart to facilitate efficient data mining of the regulatory features.
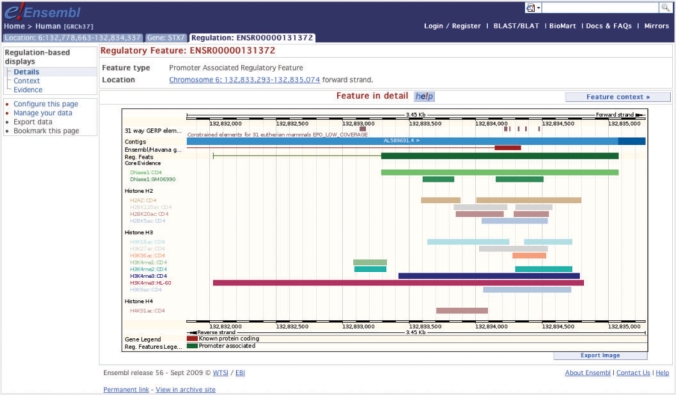
Finally, in Ensembl version 56 (September 2009), we launched a dedicated visualisation of the regulatory features in the form of a Regulation Tab at the top of the page (Figure 2). The view currently provides information about the supporting features that are used to automatically assign a preliminary regulatory function to genome regions. Our regulatory feature view will be an important area of focus over the next 12 months as we incorporate data being produced by the ENCODE project.
Figure 2.
Ensembl regulatory feature: ENSR00000131372, a promoter-associated feature located on human chromosome 6 shown with the anchoring DNase I hypersensitivity sites and supporting histone modification data.
Ensembl software and code base
As mentioned above, the Ensembl code base is being reused within the Ensembl Genomes project, which seeks to extend the Ensembl infrastructure across the taxonomic space. A number of updates to the core Ensembl infrastructure were necessary to support specific needs of Ensembl Genomes that had not been previously required by Ensembl. For example, the Ensembl core databases now support multiple species within a single core database as well as provide preliminary support for alternative transcription initiation. Support for operons is planned for the near future. The Registry component of the Ensembl API, which allows users to automatically configure database connections and other behaviours of the API, was redesigned to support connections to multiple database servers in different physical locations.
Improved data mining and analysis resources
Ensembl calculates and provides a number of key results that are useful for data integration and analyses. One of the most widely used and important examples is the identification of external references (x-refs), which was completely re-factored this year. Ensembl’s x-refs associate external database identifiers to Ensembl gene and transcript IDs and serve to enable data connections between Ensembl and biological databases such as UniProt, EMBL and RefSeq. Several x-ref assignment methods are used as described here. Direct x-refs are those where a straightforward mapping between the Ensembl ID and the external ID already exists, such as when the assignment is done by the external resource. Primary x-refs are assigned by sequence matching using Exonerate (14) between the Ensembl DNA or peptide sequences and those in the external resources. Dependent x-refs are inferred from primary x-refs where the source database references other identifiers. Finally, a class of defined priority x-refs allow for prioritisation of sources that may provide several references for the same external identifier.
We have redesigned the Ensembl Ontology database and API to make access to ontology data more consistent and straightforward. For example, Gene Ontology (GO) terms (15) and their relationships to each other are now stored in a more generic and hierarchical manner; this allows more flexible querying and the ability to perform transitive closures on GO terms which was not possible before. GO slims (http://www.geneontology.org/GO.slims.shtml) are now also supported.
The Ensembl BioMart provides access to most of our data resources in a way that facilitates the creation of complex database queries in a relatively simple manner (16). In addition to their availability through the Ensembl web site, the Ensembl BioMart is also available from the main BioMart Portal (17).
Variation
Ensembl’s variation data resources continue to be dominated by data imported from dbSNP (2). Over the past year we integrated data from the 1000 Genomes Project that was incorporated into dbSNP 130 and created initial SNP sets for orangutan and zebra finch in conjunction with the release of the gene sets for these species. In the variation web display, all SNPs are now provided with phylogenetic context if they map to a region included in one of Ensembl’s multiple alignments. The phylogenetic context includes ancestral sequence reconstructions from Ortheus (18), allowing users to look at the ancestral alleles in an evolutionary context.
By integrating data from the NHGRI curated catalogue of SNP-trait associations (19) in addition to data provided by the European Genome-phenome Archive (EGA), we have assigned annotations to over 1100 SNPs found to be associated with nearly 200 phenotypes and provided links to the published evidence. These annotations can be found on the corresponding variation page and are available through the general Ensembl search interface by searching with the phenotype name.
Web
In our last report we extensively described the fourth major design of the Ensembl web site, which was formally launched as a part of Ensembl Release 51 (November 2008) (5). With nearly 12 months of experience of the new site, we have made a number of comparably minor changes aimed at continual performance increases and reimplementation of some displays not included in the initial release of the new web code. Performance improvements included the implementation of nginx and memcached to improve server responsiveness. Visualisations reintroduced over the course of the year included multi-species comparison and alignment views that incorporate extensive contextual annotations such as genes, repeats and other features from each of the aligned species.
We have also completed major changes to the Ensembl drawing code, which allows tracks to be configured via entries in the relevant database instead of in separate static files. Finally, we have improved the ability for users to find the specific tracks that they want to display by incorporating a search box into the AJAX control panel that provides centralised page configuration.
In parallel with the new web design, we have implemented a more comprehensive monitoring of Ensembl’s performance at numerous locations around the world. We have deployed a fully functioning Ensembl mirror site to a physical location in California. This site is available at http://uswest.ensembl.org and provides an up-to-date mirror site fully monitored and maintained by Ensembl. Our tests show that users in North America and the Pacific Rim will experience faster response times from the US mirror compared to our main site in the United Kingdom and we will automatically offer users from these areas the ability to use our US mirror site as their default Ensembl. Other public Ensembl mirror sites are maintained by the user community with support from the project. Those users who take advantage of our user accounts to share settings and save sessions across multiple computers may find that the main site continues to provide faster performance due to the necessity of maintaining a single database with settings and sessions.
To address the continual growth of the size of biological databases, we have begun testing full Ensembl installations in commercial cloud computing environments. Ensembl is also currently provided as one of the free Public Data Sets on Amazon Web Services that can be integrated into any cloud based application on AWS.
Comparative genomics resources
As the number of species increases within Ensembl, our comparative genomics resources become more valuable as information sources for highly-used genomes such as human, mouse and rat. They also serve as a way to connect all aspects of the project.
One of the biggest challenges this year has been the update of the pairwise and multiple alignments to support the release of the GRCh37 human assembly. We also updated the comprehensive 31-way multi-species alignment (MSA) to include all of the low coverage mammalian genome sequences and now provide BED files for human and mouse constrained elements as determined by alignments of placental mammals. The recently published Enredo-Pecan-Ortheus (EPO) pipeline is at the heart of Ensembl’s MSA computations (18,20,21).
Ensembl GeneTrees are the result of a comprehensive analysis to predict phylogeny in vertebrates and have recently been described in detail (22). The latest improvements include the use of the meta-aligner M-Coffee (23) and incorporation of information about exon boundaries into the alignments. We now restrict our calculation of pairwise dN/dS values such that they are only calculated for high-coverage species pairs, as we found the results to be more accurate.
The current GreeTree pipeline is more robust to large gene clusters, which must be built into separate trees for computational reasons. We now annotate genes in separate trees that come from the same large cluster as distant within-species paralogues. We also annotate gene-split events (which may be real or artefactually due to an assembly problem) by analysing the protein multiple alignments: when two proteins of the same species do not overlap in the alignment, we label them the result of a gene-split event.
Visually, we added clade-specific colours to the GeneTree view to help with the interpretation of the trees. It is also possible to hide or collapse genes from pre-defined clades or from the low-coverage genomes.
Outreach and user support
Ensembl has an extensive commitment to user support, outreach and training. Provided courses include browser focused workshops introducing Ensembl to users who have never visited the site before; in depth meetings attended by developers who are building bioinformatics applications based on the Ensembl code base; and courses for clinical users interested in leveraging the Ensembl resources to help understand connections between genotype and phenotype. We also participate in regular training courses such as EBI Road Shows and Wellcome Trust Open Door Workshops that incorporate information from many of the resources developed and hosted on the Wellcome Trust Genome Campus. We aim to provide on-site training for as many of our users across the world as possible and have recently conducted trained events in Europe, North and South America, Asia, Africa and the Middle East.
We invite users interested in scheduling training to contact the Ensembl helpdesk at helpdesk@ensembl.org. For those users unable to attend a workshop in person, we are developing an extensive video library of tutorials. Our current selection is available though the Ensembl YouTube channel at http://www.youtube.com/user/EnsemblHelpdesk.
Future directions
In last year’s report, we described some of the ways that we are adapting Ensembl to the data generated by the current generation of high-throughput sequencing machines (5). We continued this theme in this report with the annotation of the first genome assembly created from combined traditional long read and next-generation short read technologies. Next year we expect to release gene sets on genome assemblies created entirely with next generation sequencing data. For a number of species, we also plan to create gene sets that incorporate short read transcriptomic data, which have shown considerable potential to increase the accuracy of our gene annotations in initial experiments using RNA-seq data from a number of zebrafish tissues.
A significant focus in the next year will be the display and annotation of variation data. Through our participation in the Locus Reference Genomic (LRG) consortium (http://www.lrg-sequence.org), we plan to incorporate summary data from Locus Specific Databases (LSDBs) at the level recommended by the community (24,25). We are also developing and testing new variation displays as part of the 1000 Genomes Project, which runs a browser based on the Ensembl code at http://browser.1000genomes.org.
FUNDING
The Ensembl project receives primary funding from the Wellcome Trust. Additional funding is provided by the European Union, BBSRC, NHGRI, NIH-NIAID and EMBL. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank all of the users of our website and other resources, and those who have provided useful feedback though our mailing list. We acknowledge those researchers, organisations and large-scale projects that have provided data to Ensembl prior to publication under the understandings of the Fort Lauderdale meeting discussing Community Resource Projects and the Toronto meeting on prepublication data sharing.
REFERENCES
- 1.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dwinell MR, Worthey EA, Shimoyama M, Bakir-Gungor B, DePons J, Laulederkind S, Lowry T, Nigram R, Petri V, et al. The Rat Genome Database 2009: variation, ontologies and pathways. Nucleic Acids Res. 2009;37:D744–D749. doi: 10.1093/nar/gkn842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.UniProt Consortium. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birney E, Andrews D, Caccamo M, Chen Y, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koscielny G, Le Texier V, Gopalakrishnan C, Kumanduri V, Riethoven JJ, Nardone F, Stanley E, Fallsehr C, Hofmann O, et al. ASTD: the Alternative Splicing and Transcript Diversity database. Genomics. 2009;93:213–220. doi: 10.1016/j.ygeno.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart–biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart Central Portal—unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paten B, Herrero J, Fitzgerald S, Beal K, Flicek P, Holmes I, Birney E. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 2008;18:1829–1843. doi: 10.1101/gr.076521.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18:1814–1828. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paten B, Herrero J, Beal K, Birney E. Sequence progressive alignment, a framework for practical large-scale probabilistic consistency alignment. Bioinformatics. 2009;25:295–301. doi: 10.1093/bioinformatics/btn630. [DOI] [PubMed] [Google Scholar]
- 22.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace IM, O'S;ullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.den Dunnen JT, Sijmons RH, Andersen PS, Vihinen M, Beckmann JS, Rossetti S, Talbot CC, Hardison RC, Povey S, Cotton RG. Sharing data between LSDBs and central repositories. Hum. Mutat. 2009;30:493–495. doi: 10.1002/humu.20977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaput J, Cotton RG, Hardman L, Watson M, Al Aqeel AI, Al-Aama JY, Al-Mulla F, Alonso S, Aretz S, et al. Planning the Human Variome Project: the Spain report. Hum. Mutat. 2009;30:496–510. doi: 10.1002/humu.20972. [DOI] [PMC free article] [PubMed] [Google Scholar]