Abstract
The Immune Epitope Database (IEDB, www.iedb.org) provides a catalog of experimentally characterized B and T cell epitopes, as well as data on Major Histocompatibility Complex (MHC) binding and MHC ligand elution experiments. The database represents the molecular structures recognized by adaptive immune receptors and the experimental contexts in which these molecules were determined to be immune epitopes. Epitopes recognized in humans, nonhuman primates, rodents, pigs, cats and all other tested species are included. Both positive and negative experimental results are captured. Over the course of 4 years, the data from 180 978 experiments were curated manually from the literature, which covers ∼99% of all publicly available information on peptide epitopes mapped in infectious agents (excluding HIV) and 93% of those mapped in allergens. In addition, data that would otherwise be unavailable to the public from 129 186 experiments were submitted directly by investigators. The curation of epitopes related to autoimmunity is expected to be completed by the end of 2010. The database can be queried by epitope structure, source organism, MHC restriction, assay type or host organism, among other criteria. The database structure, as well as its querying, browsing and reporting interfaces, was completely redesigned for the IEDB 2.0 release, which became publicly available in early 2009.
INTRODUCTION
Established in 2004 as a National Institute of Allergy and Infections Diseases (NIAID) contract, the Immune Epitope Database (IEDB) and Analysis Resource can be found at www.iedb.org. The IEDB makes information on all experimentally determined immune epitopes freely available to the public (1,2). Although, many other epitope-related databases exist (3–11), the IEDB is unique in its breadth of scope and its fine granularity. The database presents the epitopes (i.e. the molecular structures recognized by the receptors of the adaptive immune system) along with the experimental contexts in which these molecules were determined to be immune epitopes. The data are derived from all published epitope-related data available in PubMed as well as from direct submissions from scientists experimentally generating large data sets.
The scope of the database is set by the NIAID and includes, to date, epitopes related to A–C pathogens, emerging and reemerging pathogens, other infectious diseases, allergens and recently autoantigens. Currently, the database houses nearly all (92%) published experimental data related to peptidic epitopes from infectious diseases (excluding HIV) and 89% of those related to allergens. This represents an unprecedented data set, allowing end users to quickly and easily know everything that has ever been published regarding immune epitopes from a certain pathogen, in a particular host or having a particular type of experimental result. Data derived from the database can be used in various applications including, vaccine development, assay design and disease treatment.
Additionally, the database houses analysis tools related to immune epitopes (12). These tools complement the database and allow users to predict potential epitopes from antigens of interest, or to analyze known epitopes for population coverage and other relevant applications.
First available online in 2005, the IEDB recently underwent a major review resulting in significant advancements. Driven by user feedback, curator experience and the development of the Ontology of Immune Epitopes (ONTIEs) (13), data quality was scrutinized and the database schema was redesigned to accommodate new data types and features. The redesigned data schema allowed implementation of formal validation rules, which led to the identification and correction of 693 133 validation inconsistencies. This process that took ∼28 man months has resulted in drastically improved data consistency. These changes also resulted in enhanced usability for the end users and heightened the potential for future improvements. We describe here, in detail, the utility of the database as well as the advancements made as part of the IEDB 2.0 release.
Overview of the IEDB
Immune epitopes are the molecular structures recognized by adaptive immune receptors: T cell receptors (TCRs), B cell receptors (BCRs) and antibodies. The purpose of the IEDB is to catalog all experimentally derived information on immune epitopes, either as found in published manuscripts or directly submitted by the scientist generating the data. Each epitope is linked to its reference source. For published manuscripts, this information includes the authors, article title, journal name and abstract.
The types of data that are included are experiments describing recognition of an epitope by TCRs (T cell assays), BCRs or antibodies (B cell assays), as well as assays characterizing what molecular structures are presented by MHC molecules to T cells, such as elution of an epitope from an MHC molecule (MHC ligand elution assays) or experiments demonstrating the binding of an epitope to an MHC molecule (MHC binding assays). The epitope structure, source antigen and organism from which the epitope is derived are all described. The scope of the database includes data relating to epitopes derived from all infectious diseases, including NIAID Category A, B and C priority pathogens (www3.niaid.nih.gov/topics/BiodefenseRelated/Biodefense/research/CatA.html), NIAID Emerging and Reemerging infectious diseases (www3.niaid.nih.gov/topics/emerging/), allergens, and autoantigens involved in autoimmune disorders. HIV epitopes are explicitly excluded, which can instead be found in the Los Alamos HIV Molecular Immunology Database (www.hiv.lanl.gov) (14).
Rather than focusing simply on human or mouse immune responses, the IEDB does not discriminate against the hosts in which the immune response was studied, therefore all vertebrate species are potentially included.
The IEDB is supported by a contract from the NIAID and is freely available to the public. Feedback and questions are encouraged and can be sent to feedback@iedb.org.
Identifying relevant publications through PubMed
The query to extract epitope-related references consists of multiple epitope-specific keywords and logical operators. A Python script runs this query to identify all potentially relevant references catalogued in PubMed. An automated text classifier (15) is used to remove a substantial number of abstracts that are outside the scope of IEDB. This classifier is a combination of two popular machine learning methods: Naïve Bayes and support vector machine (16) and classifies each reference as either curatable or uncuratable. For each abstract that the classifier deems as curatable, the full-text manuscript is then reviewed by IEDB curators and only those still found to be curatable are entered into the database. The criteria that make a reference curatable include, but are not limited to, the presence of epitope-specific data and an explicitly defined epitope structure.
Once references have been classified as curatable, the subject matter of each publication is categorized (Table 1). Within each class, we assign a category and subcategory to further refine the topics included within the IEDB data set. The number of references present in each subcategory can be found in Supplemental Table 1. This information is not only useful to users interested in what sort of data the IEDB includes but also to scientists who may want to evaluate the relative abundance or paucity of epitope-specific data in a given research area.
Table 1.
Main categories of peptidic epitope-related references
| Main category | Total number of references | % Processed |
|---|---|---|
| Allergen | 1229 | 88.9 |
| Autoimmune | 4401 | 26.1 |
| Infectious disease | 9085 | 92.3 |
| Cancer | 2697 | 2.7 |
| HIV | 2404 | 1.4 |
| Transplant/alloantigens | 899 | 3.8 |
| Others | 3932 | 7.2 |
Curation of experimental data
Experimental data are entered by a team of PhD level curators following complex curation guidelines, established by the team with the input of immunological experts (17). These curation rules are constantly reevaluated in light of new experimental procedures and are adapted as the scope of the database grows. They are made available to the end users, in the form of a curation manual (tools.immuneepitope.org/wiki/index.php/Curation_Manual2.0), in order to ensure transparency and to encourage feedback. Each curated manuscript is reviewed for accuracy and adherence to the IEDB curation guidelines prior to becoming available to external users. Additionally, a comprehensive review of all curated data is performed prior to public release.
Acquiring data directly from investigators
In addition to published manuscripts, the IEDB also contains data that have been directly submitted by scientists. Typically, submitted data accompany the publication in which a narrower data set is described. For direct submissions, the IEDB links all data to the submitting authors, title, abstract and the date of the submission.
Currently, data submitted directly to the IEDB accounts for 33% of all epitope structures housed in the database. Data submissions can be made in XML format or by utilizing a newly developed IEDB Data Submission Tool (DST). This tool enables users to submit data utilizing tab-delimited template files which can be edited in Microsoft Excel or an equivalent application. This enhancement has significantly streamlined the submission process for users who are not familiar with the XML format. Although the submitted data are largely generated by 14 epitope discovery contract holders (18), all scientists are encouraged to contact the IEDB regarding the submission of their data. Please contact submissionsupport@iedb.org for more information.
Database design
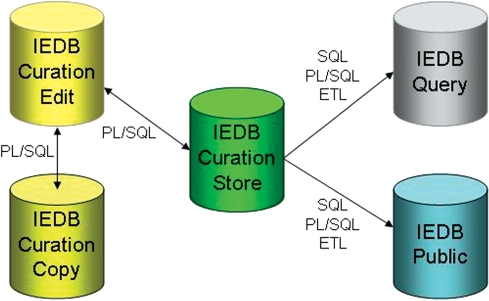
The IEDB now utilizes five distinct relational databases (Figure 1). Three of the five databases are operational databases that support the internal curation application used by the IEDB curators who enter the data. These three databases are used to store in progress and completed records and maintain the integrity of the data throughout the curation process. The remaining two databases provide data warehousing functionality and support querying, reporting and analysis activities. The IEDB Query database is designed using a dimensional approach that is optimized for querying and reporting and does not attempt to conform to any level of data normalization. The IEDB Public database, which is intended to support data analysis, is designed using a more normalized approach, though it does not achieve first normal form. Given its more flexible design, the IEDB Public database, along with its entity relationship diagram, has been made available for public download in binary and SQL formats.
Figure 1.
IEDB database architecture diagram. Data are transferred between curation databases via custom PL/SQL procedures. Data are migrated to the data warehouses via SQL and PL/SQL scripts in conjunction with an extract, transform and load (ETL) application.
IEDB ontology
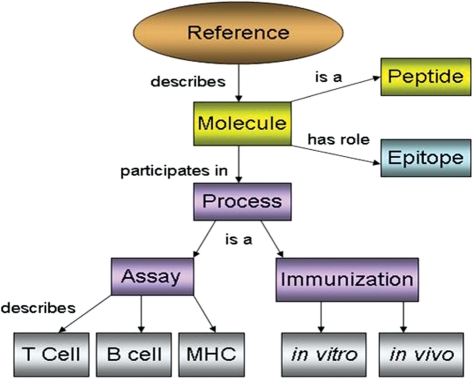
To facilitate the conversion of experimental data from text and figures in a journal publication into a computer-friendly format, we have developed the ONTIEs (ontology.iedb.org). ONTIE has been developed as a module that is imported by the Ontology for Biomedical Investigations (OBI) (The OBI Consortium http://purl.obolibrary.org/obo/obi) (19), making it immediately accessible to a wide user base. Through active ontology development, the relationships among entities stored in the database became clear. This was the major impetus for restructuring the database with a focus on objects, processes and roles as shown in Figure 2. for example, a peptide can play the role of epitope when it participates in the process of an immunological assay. The restructuring of the database around a central ‘object’ table accommodates these ontological relationships.
Figure 2.
Objects, processes and roles as represented in the IEDB.
Database content
Epitopes
Epitopes are described by two key sets of fields; those describing the molecular structure of the epitope and those describing the source from which the epitope was derived. The IEDB contains data related to linear peptide epitopes and also non-peptidic epitopes such as lipids and carbohydrates. Thus, the chemical type of the epitope is captured in the ‘Structure Type’ field. For linear peptide epitopes, by far the most commonly encountered type, the structure of that epitope is described utilizing the ‘Linear Sequence field’. The database also represents non-peptidic epitopes such as carbohydrate or lipid epitopes utilizing simplified molecular input line entry specification structures. The IEDB is currently collaborating with the Chemical Entities of Biological Interest (ChEBI) project in order to depict non-peptidic structures with proper nomenclature, synonyms and 3D structures (20).
In addition to the molecular structure of the epitope, the natural source (i.e. the ‘Source Organism’) and the molecular source (Source Antigen) of each epitope, as described by the scientist generating the data are noted. The protein sources that epitopes are derived from are described by linking to GenBank records.
The organisms from which epitopes are derived are described utilizing NCBI Taxonomy (21). The IEDB refers to the NCBI Taxonomy ID to describe an organism and also represents the organism within its place in the taxonomical tree in order to allow the end user to search via a family of organisms or on an exact strain. As part of the 2.0 release, the IEDB began supplementing the NCBI tree with strains commonly encountered in immunological research such as inbred mouse strains and viral isolates. All previously curated data were mapped to these new strains, when applicable. These supplemental organisms are also placed within the NCBI tree structure to allow the same search capabilities. As a result, users are now able to search on all laboratory animals tested in the literature.
IMMUNE RECOGNITION CONTEXT
The data housed by the IEDB is epitope-centric with every experimental assay describing the recognition of an epitope to which the assay is linked. Assays describing recognition of an epitope by TCRs (T cell assays), BCRs or antibodies (B cell assays), as well as assays characterizing MHC presentation, such as elution of an epitope from an MHC molecule (MHC ligand elution assays) or experiments demonstrating the binding of an epitope to an MHC molecule (MHC binding assays) are included in the database.
Immunization
In B cell and T cell assays, the immunization procedure whereby the host became exposed to the immunogen is captured. First, the ‘Host Organism’ in which the exposure occurred is entered by its taxonomy ID. Additional host information is captured including age, sex, disease state and disease stage at the time of the experimental assay when provided by the authors. The disease state of the host is currently described utilizing the ICD-10 classification of diseases (www.who.int/classifications/icd/en/).
Next, the ‘in vivo Process Type field’ describes how the host organism came to be exposed to the immunogen in vivo utilizing a menu of immunization types which are grouped into the following categories: occurrence of disease, administration in vivo and exposure not resulting in disease.
As part of the immunization process, the IEDB describes the immunogen to which the host was exposed along with the immunogen’s relationship to the epitope, the ‘Immunogen Epitope Relation’. For example, if the assay demonstrates the recognition of an epitope derived from influenza by antisera from an influenza-infected human, the immunogen that the host was exposed to was the influenza virus. The epitope being studied is derived from the influenza virus, thus the ‘Immunogen Epitope Relation’ is the epitope’s ‘Source Organism’. This relationship provides the end user with additional information regarding the context in which the epitope is recognized.
Additionally, the certainty of the curator that the entity being entered is the immunogen and is provided via the new ‘Immunogen Evidence Code’. This code expresses whether the author was explicit in specifying to what the host was exposed. For example when a specific strain of a virus is injected into an animal, the ‘Immunogen Evidence Code’ would be ‘Exact Match’ versus when an author is vague regarding the immunogen, as with humans exposed to an unknown strain of influenza it would be ‘Representative Selection’.
For all procedures that involved administration of the immunogen to the host, the route, dose schedule and adjuvants utilized are also captured.
The IEDB describes up to two in vivo immunizations, as is common in animals first infected with a virus and then boosted with a peptide or in humans vaccinated with a peptide vaccine and then exposed to the virus the vaccine was meant to protect against. In addition, the system can describe the in vitro immunogen in the case of primary induction of effector T cells from naïve donors or in vitro restimulation procedures performed on T cells prior to the assay.
Immunological assays
The assay types present within the IEDB reflect the assays being used in the literature. This list includes all commonly used immunological methods such as ELISA, FACs, bioassays, etc. and expands to accommodate new assays as they are developed. For all assay types, the IEDB uses an assay finder application that allows users to search on either the purpose of the assay (to measure IL-2) or the method used (ELISA). All experimental data entered into the IEDB are categorized as either positive or negative in the ‘Qualitative Measurement’ field. Additional granularity is available for positive data with values of positive-high, positive-intermediate and positive-low. For assay types with quantitative measurements, such as the KD between an antibody and epitope, the numerical values are entered along with the units. When available, the number of subjects tested, the number responding or the percent responding are also displayed.
In T cell and B cell assays, the effector cells or the antibody containing material derived from the host and being assayed is described. Effector cells are described by their tissue of origin, cell type and cell culture state at the time of the experiment. For example, blood, lymphocyte and direct ex vivo, respectively. For antibody containing material, the source of the material (for example, serum), purification state (for example, purified immunoglobulin), isotype and whether the antibodies are monoclonal or polyclonal are all captured.
In T cell assays, the antigen-presenting cells (APCs) and the restricting MHC molecule are also depicted. APCs are described similarly to the effector cells, utilizing a cell type drop down list that includes all commonly studied cell lines. The restricting MHC molecule is displayed along with an evidence code that illustrates how the restriction was determined, either in the published reference being curated or if known by the authors at the onset of the study.
For every curated experiment, there must be an assay antigen and a qualitative outcome. The assay antigen is defined as the recall antigen to which the immune response is being measured in the experimental context. The experiment may be performed in vitro (e.g. an ELISA) or in vivo (e.g. a challenge assay). As is done with the immunogen, the full description of the molecule or organism is described along with the ‘Antigen Epitope Relation’ for each antigen. Similar to the immunogen, an ‘Antigen Evidence Code’ is also used to describe how certain the curator is of the object being used as the antigen.
A new feature that was added in the 2.0 version of the IEDB is the ability to describe adoptive transfer experiments. These are procedures where the effector material (antibodies or T cells) generated in one organism (the donor) is transferred to a separate organism (the recipient) for further study. In these cases, both the donor and recipient organisms are described as mentioned for the host above. Additionally, the effector material that is transferred is explained in detail.
MHC binding assays are those in which an epitope is demonstrated to bind to a purified or cell-bound MHC molecule. MHC ligand elution assays are those that demonstrate the ability of an APC to process and present an epitope on its surface in the context of an MHC molecule. As part of the 2.0 release, all previously curated MHC molecules were edited to be represented by consistent nomenclature and reorganized by their source organism, restriction level, haplotype, class, locus, serotype and molecule name. This was a significant undertaking as the older literature used an inconsistent nomenclature. In addition, representative protein sequences were provided for alleles whenever possible. The resulting MHC allele table now provides more accurate data, more detailed information on each allele and allows for enhanced search options.
THE QUERY INTERFACE
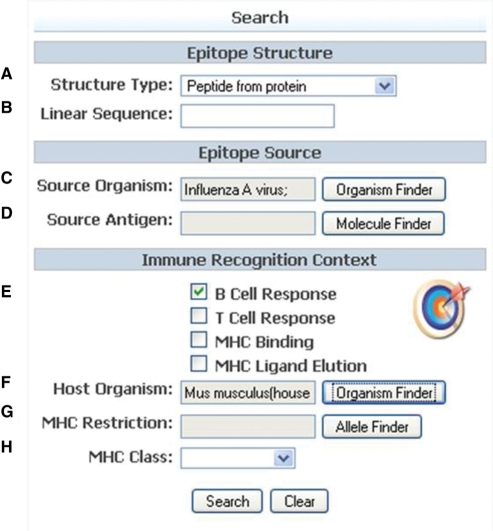
The most commonly used search functionality is now highly visible on the IEDB home page where users can directly search on criteria such as the epitope structure, source antigen, source organism or the context in which it was recognized (Figure 3). Three example queries are demonstrated in the Supplementary Data. As the same peptide may be present in many proteins from many organisms, the IEDB allows users to search on just the peptide sequence alone (Supplementary Figure 1). Additionally, data on every epitope derived from a particular protein can be queried using a protein name, organism name, chemical type or GenBank identifier (Supplementary Figure 2). All data generated on every epitope derived from a certain organism can also be retrieved (Supplementary Figure 3).
Figure 3.
Home page search. The search fields are organized into those describing the epitope structure and source (A–D) and the immune recognition context (E–H).
The search interface also allows users to limit their query to relevant immune recognition contexts (Figure 3E). For example, a user may be interested in MHC binding and T cell assays, but not B cell assays.
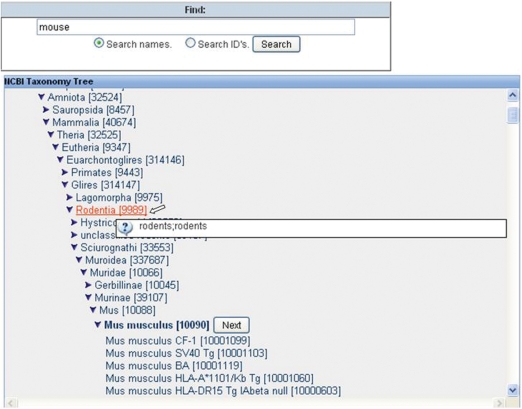
The host of the immune response (Figure 3F) may be queried by either the common or scientific name and by very narrow criteria, such as an exact strain, or by selecting any level of the NCBI tree such as all mammals, as shown in Figure 4. All common names and synonyms can be viewed by mousing over the scientific name present within the taxonomical tree.
Figure 4.
Host organism search. After entering the common name of ‘mouse’, the Organism Finder returns the scientific name of Mus musculus. The user can decide to select any higher level of the taxonomical tree, such as Rodentia (the synonym list is displayed when mousing over) or can further refine the search by selecting a specific strain.
The fields ‘MHC Restriction’ and ‘MHC Class’ (Figure 3G and H) can be used to search on the MHC molecules involved in the epitope’s recognition. One can search on all T cell data known to be restricted by a specific MHC molecule or narrow the search to look for epitopes recognized in the context of MHC Class I or MHC Class II subsets.
Table 2 demonstrates how search results are now presented in a succinct table format. Here the details of the assay are summarized by providing information on the epitope, the host of the assay, the immunization procedure, the antigen that was tested in the assay and the assay type. Every column can be sorted and all blue text or numbers are links to details pages.
Table 2.
Example assay data
| B cell ID | Reference | Epitope | Host | Immunization | Assay antigen | Antigen epitope relation | Assay description |
|---|---|---|---|---|---|---|---|
| 1498837 | J. Simeckova Rosenberg; Vaccine 1995 | KEFSEVEGRIQDLEKYV hemagglutinin HA2 (68–84) Influenza A virus | Mus musculus BALB/c | Administration in vivo with KEFSEVEGRIQDLEKYV (Epitope) | Influenza A virus | Source organism | RIA Detection of Ab/Ag binding Positive Low |
| 1501645 | J. Simeckova Rosenberg; Vaccine 1995 | KEFSEVEGRIQDLEKYV hemagglutinin HA2 (68–84) Influenza A virus | Mus musculus BALB/c | Administration in vivo with KEFSEVEGRIQDLEKYV (Epitope) | Influenza A virus | Source organism | RIA Detection of Ab/Ag binding Positive Low |
| 1583817 | M. Z. Atassi; Immunol. Commun. 1984 | KEFSEVEGRIQDLEKYV hemagglutinin HA2 (68–84) Influenza A virus | Homo sapiens | Infectious disease via exposure to Influenza A virus (Source Organism) | KEFSEVEGRIQDLEKYV hemagglutinin HA2 (68–84) Influenza A virus | Epitope | RIA Detection of Ab/Ag binding Positive Low |
| 1583818 | M. Z. Atassi; Immunol. Commun. 1984 | KEFSEVEGRIQDLEKYV hemagglutinin HA2 (68–84) Influenza A virus | Mus musculus | Administration in vivo with hemagglutinin precursor (Taxonomic Child) | KEFSEVEGRIQDLEKYV hemagglutinin HA2 (68–84) Influenza A virus | Epitope | RIA Detection of Ab/Ag binding Positive Low |
| 1583848 | M. Z. Atassi; Immunol. Commun. 1984 | KEFSEVEGRIQDLEKYV hemagglutinin HA2 (68–84) Influenza A virus | Mus musculus | Administration in vivo with KEFSEVEGRIQDLEKYV (Epitope) | KEFSEVEGRIQDLEKYV hemagglutinin HA2 (68–84) Influenza A virus | Epitope | RIA Detection of Ab/Ag binding Positive Low |
| 1583849 | M. Z. Atassi; Immunol. Commun. 1984 | KEFSEVEGRIQDLEKYV hemagglutinin HA2 (68–84) Influenza A virus | Mus musculus | Administration in vivo with KEFSEVEGRIQDLEKYV (Epitope) | Influenza A virus (A/X-31 (H3N2)) | Taxonomic child | RIA Detection of Ab/Ag binding Positive Low |
RIA, radio immuno assay. Assays are summarized by providing information on the epitope, the host of the assay, the immunization procedure, the antigen tested in the assay and the assay type. Every column header can be clicked to sort by that column. All blue text or numbers are links to further details. By clicking on the B Cell ID, one is taken to every field curated for that specific assay, which includes host demographics, administration procedures and quantitative data. The Reference link goes to every assay and epitope curated for that published manuscript or submission. The epitope link provides all information present in the database for that specific epitope, including data derived from different publications.
Advanced search functionality
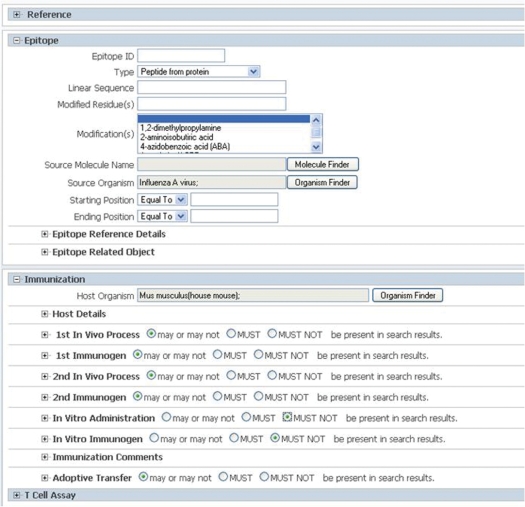
All fields of the database are searchable and can be found by utilizing the ‘Search’ dropdown menu on the home page and further refining the assay type of interest. Once in the advanced search pages, the user is shown a newly developed collapsed view of all of the IEDB fields, displaying key field groups such as Epitope, Immunization and Assay (Figure 5). Within each section, the most commonly searched fields are displayed with all fields available simply by clicking open the section. These pages are organized following the same structure as the assays themselves, for consistency, and are collapsed based upon usage, for efficiency. A complete example of a detailed search can be found in Supplementary Figure 4. The IEDB also provides tutorials on searching which can be found at tutorials.iedb.org.
Figure 5.
The advanced T cell search page. Field groups are collapsed for simplicity and can be opened or collapsed by clicking the + or − sign at each header.
Browsing by MHC allele or source organism
In addition to the simple and advanced search interfaces, the homepage provides a keyword search option and ‘browse by’ links, either by MHC allele or by source organism. When browsing by MHC allele, one is shown the currently curated data on MHC alleles organized utilizing the NCBI taxonomy and a hierarchy of MHC types. For example, if one is interested in human alleles and clicks on the Homo sapiens icon, the number of Class I, Class II and non-classical structures for which data exists are displayed. Within each category, the data are further organized by their locus, serotype and molecule. When browsing by source organism, the data are organized following the NCBI taxonomy and allows the user to search for a particular organism by either common or scientific name. Once a search result has been found, the user has the option of directly viewing the data related to their search or to view their results within the taxonomical tree, allowing one to either broaden the search to a higher node or to refine it by selecting a specific strain. In addition, the number of epitopes derived from each source organism is provided in the tree to help the user better identify relevant results.
DATA AVAILABILITY
The IEDB is found at www.iedb.org and is organized into three sections; query interface, up to date metrics on database content and links to resources.
The IEDB is freely available online and allows users to export their search results in Microsoft Excel format. The data can be exported in either a compact or detailed format. Additionally, users may also download an export of the schema, tables or the entire database in either XML or MySQL formats.
SUPPORT
At the new Solutions Center (iedb.zendesk.com/portal), one will find online and downloadable documents describing the database, the new features of the 2.0 release, the IEDB monthly newsletter and forums discussing topics that end users have asked about. In addition, the site provides users with detailed information on how the IEDB staff curates references and how to query the database. A shorter version of the curation manual, tailored for the database user, is provided at tools.immuneepitope.org/wiki/index.php/Data_Field_Descriptions. Links to provide feedback, questions and suggestions are also present (help@iedb.org).
OUTLOOK FOR THE IEDB AND ITS USERS
The content of the IEDB is current regarding all published peptidic epitope data relating to infectious disease and allergens. Within the next year, we expect to complete the curation of autoimmune-related epitopes and to bring non-peptidic epitope data into the database. In order to remain current on new publications, the IEDB reruns the PubMed query quarterly and adds all new relevant publications. New data submissions are ongoing and we expect to increase their number as NIAID contracts requiring data submission are fulfilled. Additionally, the new DST affords greater access to the submission process for all scientists wishing to deposit their data in the IEDB. As new data are entered, the IEDB continues to adapt data fields, reevaluate curation guidelines and seek feedback from end users.
Immunology is a constantly expanding field and the database and ontology will continue to evolve in parallel as new assays are developed or new nomenclature is implemented. The IEDB will continue to expand its collaboration with other resources. The ability to link to other databases was implemented as part of the 2.0 release, allowing external databases to directly link to relevant information present on our web site. We hope this feature will become widely used as we continue to reach out to new resources.
The IEDB will continue solicitation of feedback from users in order to continue improving all aspects of the database. We are eager for scientists to visit iedb.org and let us know what you think help@iedb.org.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health/National Institute of Allergy and Infections Diseases, Immune Epitope Database (contract number: HHSN2662004000 0 6C), under the Immune Epitope Database and Analysis Program.
Conflicts of interest statement. None declared.
REFERENCES
- 1.Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters B, Sette A. Integrating epitope data into the emerging web of biomedical knowledge resources. Nat. Rev. Immunol. 2007;7:485–490. doi: 10.1038/nri2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toseland CP, Clayton DJ, McSparron H, Hemsley SL, Blythe MJ, Paine K, Doytchinova A, I, Guan P, Hattotuwagama CK, Flower DR. AntiJen: a quantitative immunology database integrating functional, thermodynamic, kinetic, biophysical, and cellular data. Immunome Res. 2005;1:4. doi: 10.1186/1745-7580-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blythe MJ, Doytchinova IA, Flower DR. JenPep: a database of quantitative functional peptide data for immunology. Bioinformatics. 2002;18:434–439. doi: 10.1093/bioinformatics/18.3.434. [DOI] [PubMed] [Google Scholar]
- 5.Saha S, Bhasin M, Raghava GPS. Bcipep: a database of B-cell epitopes. BMC Genomics. 2005;6:79. doi: 10.1186/1471-2164-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlessinger A, Ofran Y, Yachdav G, Rost B. Epitome: database of structure-inferred antigenic epitopes. Nucleic Acids Res. 2006;34:D777–D780. doi: 10.1093/nar/gkj053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh MK, Srivastava S, Raghava GP, Varshney GC. HaptenDB: a comprehensive database of haptens, carrier proteins and anti-hapten antibodies. Bioinformatics. 2006;22:253–255. doi: 10.1093/bioinformatics/bti692. [DOI] [PubMed] [Google Scholar]
- 8.Yang IS, Lee JY, Lee JS, Mitchell WP, Oh HB, Kang C, Kim KH. Influenza sequence and epitope database. Nucleic Acids Res. 2008;37:D423–D430. doi: 10.1093/nar/gkn881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govindarajan KR, Kangueane P, Tan TW, Ranganathan S. MPID: MHC-Peptide Interaction Database for sequence-structure-function information on peptides binding to MHC molecules. Bioinformatics. 2003;19:309–310. doi: 10.1093/bioinformatics/19.2.309. [DOI] [PubMed] [Google Scholar]
- 10.Lata S, Bhasin M, Raghava GP. MHCBN 4.0: a database of MHC/TAP binding peptides and T-cell epitopes. BMC Res. Notes. 2009;20:61. doi: 10.1186/1756-0500-2-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Günther S, Hempel D, Dunkel M, Rother K, Preissner R. SuperHapten: a comprehensive database for small immunogenic compounds. Nucleic Acids Res. 2007;35:D906–D910. doi: 10.1093/nar/gkl849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, Bui HH, Buus S, Frankild S, Greenbaum J, et al. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenbaum JA, Vita R, Zarebski L, Emami H, Sette A, Ruttenberg A, Peters B. Proceedings of the 12th Annual Bio-Ontologies Meeting. International Society for Computational Biology. 2009. Representing the Immune Epitope Database in OWL; pp. 45–48. [Google Scholar]
- 14.Korber BTM, Brander C, Haynes BF, Koup R, Moore JP, Walker BD, Watkins DI. Los Alamos, New Mexico: Los Alamos National Laboratory, Theoretical Biology and Biophysics; 2007. HIV Molecular Immunology 2006/2007. [Google Scholar]
- 15.Wang P, Morgan AA, Zhang Q, Sette A, Peters B. Automating document classification for the Immune Epitope Database. BMC Bioinformatics. 2007;26:269–279. doi: 10.1186/1471-2105-8-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joachims T. Dissertation. Kluwer: The Netherlands; 2002. Learning to Classify Text Using Support Vector Machines. [Google Scholar]
- 17.Vita R, Peters B, Sette A. The curation guidelines of the immune epitope database and analysis resource. Cytometry A. 2008;73:1066–1070. doi: 10.1002/cyto.a.20585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sette A, Fleri W, Peters B, Sathiamurthy M, Bui HH, Wilson S. A roadmap for the immunomics of category A-C pathogens. Immunity. 2005;22:155–161. doi: 10.1016/j.immuni.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 19.The OBI Consortium. Proceedings of the 12th Annual Bio-Ontologies 35 Meeting. International Society for Computational Biology. 2009. Modeling biomedical experimental processes with OBI; pp. 41–44. [Google Scholar]
- 20.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The IEDB is found at www.iedb.org and is organized into three sections; query interface, up to date metrics on database content and links to resources.
The IEDB is freely available online and allows users to export their search results in Microsoft Excel format. The data can be exported in either a compact or detailed format. Additionally, users may also download an export of the schema, tables or the entire database in either XML or MySQL formats.