Abstract
Although protein–peptide interactions are estimated to constitute up to 40% of all protein interactions, relatively little information is available for the structural details of these interactions. Peptide-mediated interactions are a prime target for drug design because they are predominantly present in signaling and regulatory networks. A reliable data set of nonredundant protein–peptide complexes is indispensable as a basis for modeling and design, but current data sets for protein–peptide interactions are often biased towards specific types of interactions or are limited to interactions with small ligands. In PepX (http://pepx.switchlab.org), we have designed an unbiased and exhaustive data set of all protein–peptide complexes available in the Protein Data Bank with peptide lengths up to 35 residues. In addition, these complexes have been clustered based on their binding interfaces rather than sequence homology, providing a set of structurally diverse protein–peptide interactions. The final data set contains 505 unique protein–peptide interface clusters from 1431 complexes. Thorough annotation of each complex with both biological and structural information facilitates searching for and browsing through individual complexes and clusters. Moreover, we provide an additional source of data for peptide design by annotating peptides with naturally occurring backbone variations using fragment clusters from the BriX database.
INTRODUCTION
A growing number of interactions are known to be mediated by short linear peptides (1). It is estimated that 15–40% of all interactions in the cell are protein–peptide interactions (2,3), which indicates that a large portion of the proteome is either directly or indirectly involved in peptide-binding events. Peptide-mediated interactions are normally short-lived and therefore found most in signaling and regulatory networks where fast response to stimuli is required (4). Many databases have been implemented that assemble the sequence patterns involved in such interactions, such as the Eukaryotic Linear Motif (ELM) database (5), PROSITE (6) and SCANSITE (7).
Unfortunately, the estimated abundance of protein–peptide interactions from the genome is not reflected in the number of available 3D protein–peptide complexes. While many protein–protein and protein–domain interaction databases with structural annotations exist (8–12), only few of them explicitly consider protein–peptide interactions (13). Moreover, focus on specific types of peptide interactions (PDZ domains, SH3 domains) has biased the content of structural databases. Grouping of 3D structures of protein–peptide complexes into functional modules has been established by several methods, such as using ELM patterns [e.g. 3did (13)] and multiple sequence alignment of the ligands [e.g. FireDB (14)]. Additionally, specialized databases focusing on a specific functional group have been published, such as PROCOGNATE for enzyme complexes (15), MPID-T for T-cell receptors (16) and the HMRBase for hormone-receptor data (17). For a detailed list with related databases we refer to Supplementary Table S1. In contrast, our objective was to build an unbiased collection of nonredundant peptide binding sites, where grouping is based solely on 3D similarity and no bias for functional relations or sequence similarity is introduced.
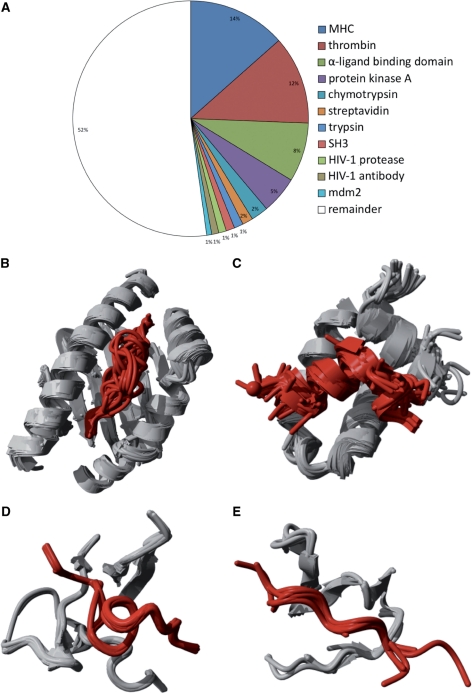
To this end, we have mined the Brookhaven Protein Data Bank (PDB) for protein–peptide complexes using rigid quality parameters, and thus obtained 1431 high-resolution 3D structures (see Methods section for details on the selection procedure). These complexes were clustered based on 3D similarity into 505 unique protein–peptide interface clusters, representing the full structural diversity of protein–peptide complexes available in the PDB. The aforementioned bias for specific peptide interactions is demonstrated in the further clustering of these complexes. Of all protein–peptide complexes available from the PDB, 47% are clustered within only 10 classes (Figure 1A), containing complexes with peptides bound to Major Histocompatibility Complex (MHC) (14%, Figure 1B), thrombins (12%, Figure 1D), α-ligand binding domains (8%, Figure 1C), protein kinase A, chymotrypsin, streptavidin, trypsin, SH3 domains (Figure 1E), HIV-1 protease, HIV-1 antibody and mdm2.
Figure 1.
Contents of the protein–peptide dataset. From the PDB, 1431 protein–peptide interactions are extracted and clustered using the architecture of the binding site to remove redundancy. Of all the protein–peptide complexes, 47% are classified in 10 classes with more than five members, while the remaining 53% contain less frequent structural binding modes (A). Clusters with (B) class I MHC bound to peptide (169 structures), (C) estrogen receptor α-ligand binding domain bound to peptide (111 structures), (D) thrombin inhibitor complex (89 structures) and (E) SH3 domain–peptide interaction (7 structures) are shown.
DATABASE CONTENTS
Construction of a nonredundant dataset of protein–peptide complexes
We have filtered the Brookhaven Protein Data Bank (PDB) (18) for protein–peptide complexes requiring X-ray structures with a resolution lower than 2.5 Å, peptides with a size from 5 to 35 amino acids, peptides containing natural amino acids only, receptors with a minimum size of 35 amino acids, and the first unit in the PDB in case of crystallographic symmetry. As a result, 1431 complexes were retained and clustered on their binding architecture using an adaptation of the Hierarchical Agglomeration algorithm used for constructing BriX (19), a database of protein fragments. RMSD between any two complexes superposed on backbone Cα atoms has been computed using MUSTANG to allow for structural alignment of unrelated protein structures (20). Any two structures are grouped together if they superpose below 2 Å RMSD for at least 75% of their interfaces. In this way, we retained 505 unique protein–peptide interface clusters. Furthermore, we clustered the protein–peptide complexes using RMSD values of 1, 2 and 3 Å combined with structural alignment of 50%, 75% and 95% of the interfaces, respectively. The clusters vary slightly depending on those parameters. The distribution of the number of elements in the PepX clusters for various thresholds of structural similarity (Ångstrom) and structural alignment of the binding site is shown in Supplementary Figure S1. For all settings most clusters contain only one complex: 64% of all clusters are singletons for thresholds of 3 Å and 50% alignment (Supplementary Figure S1A), whereas 87% of all clusters resulting from 1 Å RMSD and 95% alignment (Supplementary Figure S1C) contain only one element.
PepX statistics
The upper threshold for the peptide length was set to 35 amino acids, but the majority of the peptides are between 5 and 15 residues long, with a peak at 9 residues (Supplementary Figure S2). The size of receptors varies between 67 and 7073 residues, and the largest fraction lies in the [400–500] range (Supplementary Figure S3).
The receptor sequences in the PepX database were clustered with the cd-hit algorithm (21) for various thresholds, resulting in datasets where sequences with 40–100% sequence identity are removed (Supplementary Figure S4). Although there is large sequence redundancy within the database (removing sequences with >40% sequence identity results in removing >70% of all complexes in the database), this does not always reflect a redundancy in binding modes. For instance, MHCs have high-sequence identity but bind a wide range of peptides in different modes (22,23). Preliminary analysis of the sequence redundancy in the full complex dataset versus the dataset with cluster centroids revealed that using geometric properties for clustering removes most sequence identity without discarding relevant structural binding motifs.
All receptors in protein–peptide complexes have been annotated with the structural classifications SCOP (24) and CATH (25) based on the PDB ID and chain of the receptor (18) and with PFAM (26) based on the UniProt identifier (27). The coverage of PepX is highest for UniProt (82%), followed by structural classifications by CATH (71%) and SCOP (56%), and finally protein family annotation by Pfam (50%) (Supplementary Figure S4). Within these annotations, we have analyzed in detail the occurrence of PepX complexes in the various levels of the structural hierarchies represented in SCOP and CATH. Although most SCOP classes are represented by receptors in the database, protein–peptide complexes do not represent the full range of SCOP folds (8%), superfamilies (6%) and families (4%) (Supplementary Figure S6). When we look at the distribution of receptors in the different SCOP classes with respect to the distribution of PDB structures in the full SCOP database (Supplementary Figure S7), we see that in PepX the all-β and α + β classes are clearly overrepresented (30 versus 24% for the all-β class, 38 versus 25% for the α + β class, respectively). Similar results are obtained for the CATH classifications: the complexes represent every CATH class, and architectures are highly represented as well (Supplementary Figure S8). In contrast, at lower CATH levels, <10% of both topologies and superfamilies hold at least one protein–peptide complex. In accordance with the SCOP analysis, classes with mainly β-structures are largely overrepresented in PepX (Supplementary Figure S9). Alpha and beta structures are underrepresented (35% in PepX versus 52% full CATH). This is also seen in SCOP when we merge the classes together (α/β and α + β), although the difference is smaller (43% PepX versus 49% full SCOP).
Ligand annotation with structural variants for peptide design
Given the scarcity of protein–peptide structures and their obvious relevance in drug design (28–32), we provide an additional service for peptide design. Since it was recently shown that protein–peptide interactions can be reliably mimicked using interacting fragments from monomeric proteins (33), it is possible to provide structural variations of peptide ligands using protein fragments. Each ligand peptide in the PepX dataset is associated with its corresponding structural class from the database of protein fragment classes, BriX (http://brix.vub.ac.be) (19). Sets of protein fragments with highly similar backbone structure are grouped in these fragment classes. Each protein fragment class represents a natural variation on a typical backbone conformation. Mapped on protein–peptide pairs, these structural classes can be used to model and design alternative peptides with slightly adapted backbone conformation that better fit given amino acid sequences.
Database availability
PepX is accessible through a web portal at http://pepx.switchlab.org. The full database with annotations is available for download both in SQL format and as flat files. The entire dataset of 1431 PDBs with binding site residues and the equivalent centroid dataset of 505 binding sites can be downloaded. PepX is monthly updated with new 3D structures from the PDB. The PepX web server is implemented using the Drupal Content Management system (http://www.drupal.org). Images of the 3D structures were generated using the Yasara tool suite (http://www.yasara.org).
DATABASE ACCESS
User interface
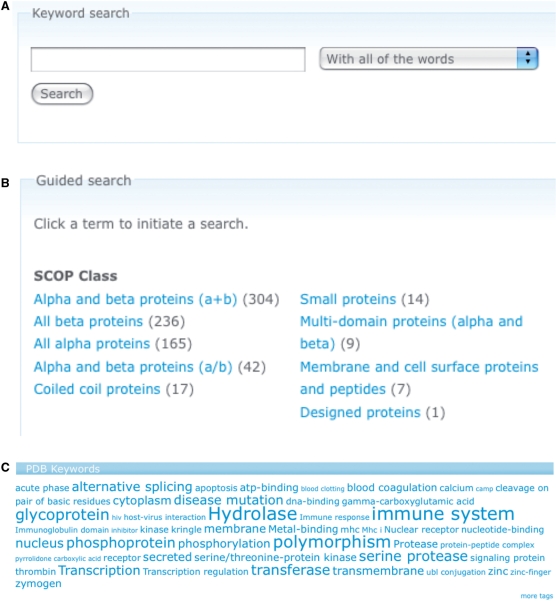
Extensive search and browse facilities are implemented for the PepX web site. Browsing the database can be performed at two levels: individual complex structures and clusters of complexes. In the latter case, the user can choose the level of similarity within one cluster by adjusting the root mean square distance between structures within one cluster and the percentage of structural alignment between binding sites. The full PepX database can be searched through a simple Google-like search box, which uses a full index of all information contained in the database (Figure 2A). The guided search allows searching the database in specific subgroups, generated from the structural classifications and keywords (Figure 2B). In addition, tag clouds of the structural annotations can be used to generate specialized listings of protein–peptide complexes (Figure 2C).
Figure 2.
Search options in the PepX database. (A) A simple, Google-like search on the contents of the database is implemented. The search is nonrestrictive and accepts everything from keywords to PDB identifiers. (B) Guided search uses structural classifications of SCOP and CATH and keywords from PDB and Pfam. (C) Tag clouds are generated from the various annotations of the protein–peptide complexes.
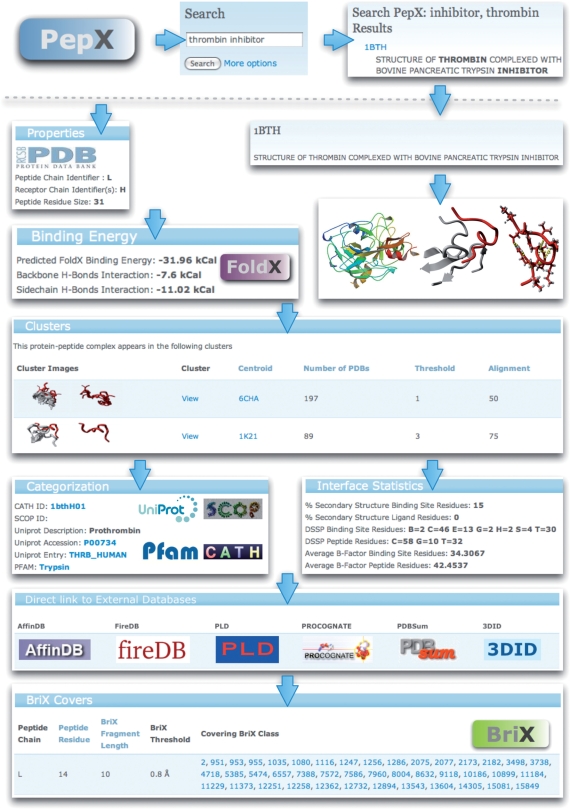
For each individual complex, several types of information are shown (Figure 3). Besides general information of the complex (PDB ID, chains), functional and structural annotation of the protein (UniProt, SCOP, CATH), also detailed structural information about the interaction itself is displayed. The binding affinity for the protein–peptide complex is calculated using the FoldX force field (34) and details of the contribution of backbone and side chain hydrogen bonds as well as the total binding energy is shown. The binding site is structurally characterized using several metrics such as secondary structure content, and 3D images of the binding site and the ligand itself were generated to illustrate the specific parts of the protein contributing to the binding site. Furthermore, all the clusters the complex takes part in are listed. Clicking on a specific cluster reveals a detailed page containing information on the centroid complex of the cluster as well as the list of all complexes belonging to the cluster.
Figure 3.
Overview of the information displayed for a thrombin complexed with an inhibitor. Searching for the keywords ‘thrombin’ and ‘inhibitor’ provides a list of hits. For the selected entry 1BTH various types of information are shown, as well as a listing of the clusters the complex belongs to. General properties of the PDB entry are accompanied by 3D views of the full complex and detailed views of the peptide-binding site generated by Yasara. The binding energy between protein and peptide as calculated by the FoldX force field is shown together with details for the hydrogen bond interactions. Various statistics regarding the secondary structure content and flexibility parameters for the binding site are listed, as well as direct links to relevant databases. The peptides are annotated with naturally occurring backbone variations using fragment clusters from the BriX database.
Automated database interaction through web-based API
All information contained within the PepX database is exposed as XML (extensible markup language). When certain URLs are visited, an XML file with the requested data is returned, following the REST interface for data exchange. For example, calling the URL http://pepx.switchlab.org/clusters.xml?threshold=2&alignment=75 serves an XML file with a description of the clusters for threshold 2 Å and an alignment of 75%. The XML interface is implemented for clusters, PDBs and BriX classes providing backbone variations on the peptides.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
PhD scholarship from the Institute for Science and Innovation Flanders (IWT) (to P.V. and L.B.); Institute for the encouragement of Scientific Research and Innovation of Brussels (ISRIB) (to J.R.); EC projects: 3D repertoire (EC 512028) and Prospects (in part). Funding for open access: PhD scholarship from the Institute for Science and Innovation Flanders (IWT) (to P.V. and L.B.); Institute for the encouragement of Scientific Research and Innovation of Brussels (ISRIB) (to J.R.); EC projects: 3D repertoire (EC 512028) and Prospects (in part).
Conflict of interest statement. None declared.
REFERENCES
- 1.Neduva V, Russell RB. Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 2006;17:465–471. doi: 10.1016/j.copbio.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Neduva V, Linding R, Su-Angrand I, Stark A, de Masi F, Gibson TJ, Lewis J, Serrano L, Russell RB. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petsalaki E, Russell RB. Peptide-mediated interactions in biological systems: new discoveries and applications. Curr. Opin. Biotechnol. 2008;19:344–350. doi: 10.1016/j.copbio.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 5.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghavachari B, Tasneem A, Przytycka TM, Jothi R. DOMINE: a database of protein domain interactions. Nucleic Acids Res. 2008;36:D656–D661. doi: 10.1093/nar/gkm761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogmen U, Keskin O, Aytuna AS, Nussinov R, Gursoy A. PRISM: protein interactions by structural matching. Nucleic Acids Res. 2005;33:W331–W336. doi: 10.1093/nar/gki585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong S, Yoon G, Jang I, Bolser D, Dafas P, Schroeder M, Choi H, Cho Y, Han K, Lee S, et al. PSIbase: a database of Protein Structural Interactome map (PSIMAP) Bioinformatics. 2005;21:2541–2543. doi: 10.1093/bioinformatics/bti366. [DOI] [PubMed] [Google Scholar]
- 12.Chen YC, Lo YS, Hsu WC, Yang JM. 3D-partner: a web server to infer interacting partners and binding models. Nucleic Acids Res. 2007;35:W561–W567. doi: 10.1093/nar/gkm346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein A, Panjkovich A, Aloy P. 3did Update: domain–domain and peptide-mediated interactions of known 3D structure. Nucleic Acids Res. 2009;37:D300–D304. doi: 10.1093/nar/gkn690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez G, Valencia A, Tress M. FireDB–a database of functionally important residues from proteins of known structure. Nucleic Acids Res. 2007;35:D219–D223. doi: 10.1093/nar/gkl897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bashton M, Nobeli I, Thornton JM. PROCOGNATE: a cognate ligand domain mapping for enzymes. Nucleic Acids Res. 2008;36:D618–D622. doi: 10.1093/nar/gkm611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong JC, Kong L, Tan TW, Ranganathan S. MPID-T: database for sequence-structure-function information on T-cell receptor/peptide/MHC interactions. Appl. Bioinformatics. 2006;5:111–114. doi: 10.2165/00822942-200605020-00005. [DOI] [PubMed] [Google Scholar]
- 17.Rashid M, Singla D, Sharma A, Kumar M, Raghava GP. Hmrbase: a database of hormones and their receptors. BMC Genomics. 2009;10:307. doi: 10.1186/1471-2164-10-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kouranov A, Xie L, de la Cruz J, Chen L, Westbrook J, Bourne PE, Berman HM. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baeten L, Reumers J, Tur V, Stricher F, Lenaerts T, Serrano L, Rousseau F, Schymkowitz J. Reconstruction of protein backbones from the BriX collection of canonical protein fragments. PLoS Comput. Biol. 2008;4:e1000083. doi: 10.1371/journal.pcbi.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 22.Collins EJ, Garboczi DN, Wiley DC. Three-dimensional structure of a peptide extending from one end of a class I MHC binding site. Nature. 1994;371:626–629. doi: 10.1038/371626a0. [DOI] [PubMed] [Google Scholar]
- 23.Elliott T, Neefjes J. The complex route to MHC class I-peptide complexes. Cell. 2006;127:249–251. doi: 10.1016/j.cell.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuff AL, Sillitoe I, Lewis T, Redfern OC, Garratt R, Thornton J, Orengo CA. The CATH classification revisited–architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium U. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parthasarathi L, Casey F, Stein A, Aloy P, Shields DC. Approved drug mimics of short peptide ligands from protein interaction motifs. J. Chem. Inform. model. 2008;48:1943–1948. doi: 10.1021/ci800174c. [DOI] [PubMed] [Google Scholar]
- 29.Van Der Sloot A, Kiel C, Serrano L, Stricher F. Protein design in biological networks: from manipulating the input to modifying the output. Protein Eng. Des. Select. 2009:1–6. doi: 10.1093/protein/gzp032. [DOI] [PubMed] [Google Scholar]
- 30.Reina J, Lacroix E, Hobson SD, Fernandez-Ballester G, Rybin V, Schwab MS, Serrano L, Gonzalez C. Computer-aided design of a PDZ domain to recognize new target sequences. Nat. Struct. Biol. 2002;9:621–627. doi: 10.1038/nsb815. [DOI] [PubMed] [Google Scholar]
- 31.Yin H, Slusky J, Berger B, Walters R, Vilaire G, Litvinov R, Lear J, Caputo G, Bennett J, Degrado W. Computational design of peptides that target transmembrane helices. Science. 2007;315:1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 32.Ballinger MD, Shyamala V, Forrest LD, Deuter-Reinhard M, Doyle LV, Wang JX, Panganiban-Lustan L, Stratton JR, Apell G, Winter JA, et al. Semirational design of a potent, artificial agonist of fibroblast growth factor receptors. Nat. Biotechnol. 1999;17:1199–1204. doi: 10.1038/70746. [DOI] [PubMed] [Google Scholar]
- 33.Vanhee P, Stricher F, Baeten L, Verschueren E, Lenaerts T, Serrano L, Rousseau F, Schymkowitz J. Protein–peptide interactions adopt the same structural motifs as monomeric protein folds. Structure. 2009;17:1128–1136. doi: 10.1016/j.str.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 34.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acid Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]