Abstract
Cancer-related investigations have long been in the limelight of biomedical research. Years of effort from scientists and doctors worldwide have generated large amounts of data at the genome, transcriptome, proteome and even metabolome level, and DNA and RNA cancer signature databases have been established. Here we present a database of differentially expressed proteins in human cancers (dbDEPC), with the goal of collecting curated cancer proteomics data, providing a resource for information on protein-level expression changes, and exploring protein profile differences among different cancers. dbDEPC currently contains 1803 proteins differentially expressed in 15 cancers, curated from 65 mass spectrometry (MS) experiments in peer-reviewed publications. In addition to MS experiments, low-throughput experiment data from the same literatures and cancer-associated genes from external databases were also integrated to provide some validation information. Furthermore, dbDEPC associates differential proteins with important structural variations in the human genome, such as copy number variations or single nucleotide polymorphisms, which might be helpful for explaining changes in protein expression at the DNA level. Data in dbDEPC can be queried by protein identifier, description or sequence; the retrieved protein entry provides the differential expression pattern seen in cancers, along with detailed annotations. dbDEPC is expected to be a reference database for cancer signatures at the protein level. This database is provided at http://dbdepc.biosino.org/index/.
INTRODUCTION
Cancer is a class of diseases in which abnormal cells divide without control and are able to invade other tissues (1). These diseases are a leading cause of death worldwide, accounting for seven million deaths in 2008 (based on ‘World Cancer Report 2008’, from the WHO). A promising approach to cancer detection and treatment is based on reliable biomarker discovery, which assesses molecular changes between cancerous and normal tissue. For many years, scientists have been searching for biomarkers to aid in cancer diagnosis, prognosis and therapy (2). Knowledge of cancer biomarkers has increased greatly with improvements in screening methods, from conventional experiments (e.g. gel electrophoresis and immunohistochemistry) to high-throughput technologies (e.g. microarray, mass spectrometry [MS] and sequencing).
In recent decades, several cancer-related database systems have been developed to associate human genes with cancers. CGED (3) and ITTACA (4) collect cancer-related gene expression and clinical data. TAG (http://www.binfo.ncku.edu.tw/TAG/GeneDoc.php, 519genes) and TGDBs (http://www.tumor-gene.org/Breast/index.html, 300genes) provide repositories for cancer-related genes (e.g. oncogenes, tumor suppressor genes). COSMIC (5) stores 78 933 somatic mutation information related to human cancers. All these databases focus on DNA/RNA level changes, such as mutation, single nucleotide polymorphisms (SNPs) and mRNA expression change. Most pathophysiologic changes in cancer, however, are mediated by expression change at the protein level (6).
In the post-genomic era, proteomics has been developed to quantify protein expression changes. The primary technology of quantitative proteomics is stable isotope labeling combined with liquid chromatography tandem MS (LC-MS/MS) (7), which has been used successfully in lung cancer (8–10), breast cancer (11–13), leukemia (14) and other cancer types. For example, Ralhan et al. employed iTRAQ-2DLC-MS/MS to identify potential biomarkers of oral premalignant lesions (OPLs), and the subsequent immunohistochemistry verified that the combination of three best-performing biomarkers achieved a sensitivity of 0.83 and a specificity of 0.74 in discriminating OPLs from normal oral tissues (15).
With more and more cancer proteomic research being performed, a unifying bioinformatic resource is required to collect and analyze the resulting data. Thus we have developed a manually curated database of differentially expressed proteins in human cancers (dbDEPC). This database integrates results from various MS experiments to provide protein-level expression changes for 1803 proteins in 15 cancers. Compared with previous cancer-related databases, dbDEPC is a novel resource, in providing information on differentially expressed proteins; thus, it may become an important repository for candidate protein biomarkers. Genomic structure variations also are provided in dbDEPC, to combine information on cancer-related protein-level and DNA-level changes. All data can be searched, browsed and downloaded directly from the website.
DATA COLLECTION
A semi-automatic method was used to curate differentially expressed proteins from peer-reviewed MS-related papers, as follows:
PubMed was automatically searched using MS-related keywords (mass spectrometry, proteomics, quantitative) and cancer-related keywords (cancer, carcinoma), with search results limited to papers published before April 2009 for the current version of dbDEPC.
To control data quality, retrieved papers were selected by manually checking whether protein identification and differential expression filtering were implemented by strict score cutoff or P-value of a statistical test. After this quality control step, 65 MS experiments in 47 papers were retained for further data collection (Supplementary Table S1). Differentially expressed proteins had ≥1.5-fold change in 63% of experiments and P < 0.05 in 25% of experiments, confirming high data quality. Among 65 experiments, 58 experiments were designed for human cancers, 5 for mouse and 2 for rat. Although dbDEPC focuses on differentially expressed proteins in human cancers, proteins in mouse and rat were kept for future development.
The 47 original papers retained in step (ii) used protein ID numbers from various versions of different databases. Based on these ID numbers, we retrieved each primary sequence from the appropriate database and mapped sequences to the uniform IPI database (16) (human IPI version 3.60, mouse and rat IPI version 3.56), using the BLASTP program (17) (the e-value cutoff was set to 10−8, the BLAST-HSP coverage was >0.95).
In addition to MS experiments, low-throughput (LTP) experiments, including western blot, immunohistochemistry, ELISA and real-time PCR, often were implemented in original publications to validate the differential expression of potential biomarkers. Such validation information was imported into dbDEPC.
The PAnnBuilder package (18) was utilized to acquire general protein annotation information, such as protein description, gene symbol, Entrez gene ID, Gene Ontology (19), Pfam domain (20) and associated pathways (21). Additionally, SNPs and other genomic variations were downloaded from the dbSNP (22) and DGV databases (23), and integrated into dbDEPC.
To associate dbDEPC with other cancer-related databases, CGDCP and CGC were integrated into dbDEPC by Entrez gene ID. [CGDCP is the Cancer Gene Data Curation Project, which associates genes and diseases by text mining and manual checking (http://ncicb.nci.nih.gov/projects/cgdcp); CGC is the Cancer Gene Census, which collects cancer-related gene mutations (http://www.sanger.ac.uk/genetics/CGP/Census/).]
DATABASE CONSTRUCTION
dbDEPC consists of a relational database and a dynamic web interface, constructed in the Python 2.6 programming language (http://www.python.org/), configured on a RedHat Linux server, and run via a Django web framework (http://www.djangoproject.com/) with an Apache server (http://www.apache.org/).
DATABASE CONTENT
Currently, dbDEPC contains 1803 differentially expressed proteins from 15 cancers: lung adenocarcinoma, gastric cancer, hepatocellular carcinoma (HCC), colorectal cancer, breast cancer, prostate cancer, esophageal cancer, cervical cancer, pancreatic carcinoma, ovarian cancer, leukemia, thyroid cancer, OPLs, uterine cancer and oral cancer. A few proteins in mouse and rat are included in dbDEPC, but we focus on 1487 human proteins (1370 human genes) in this paper.
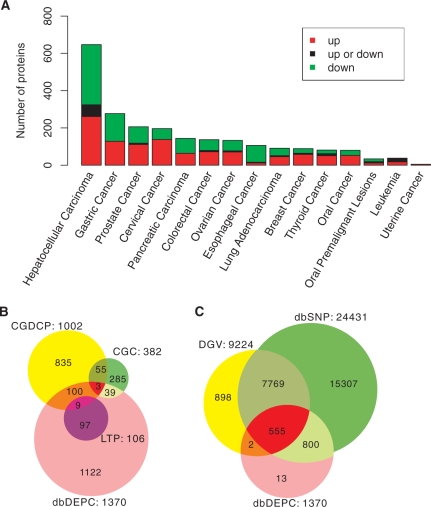
Figure 1A illustrates the number of differentially expressed proteins collected for each cancer type. Taking HCC as an example, 647 proteins are differentially expressed, among which 261 proteins are up-regulated and 323 proteins are down-regulated. Another 63 proteins show conflicting expression change (up or down) in different HCC experiments, which might be due to different HCC samples or experimental protocols. dbDEPC provides more informative and reliable data than does a single experiment under such circumstances, showing the advantage of data integration.
Figure 1.
Data content of dbDEPC. (A) Number of differentially expressed human proteins in each cancer. (B) Overlap between dbDEPC and two external databases. (C) Statistics for genomic variations in human genes collected in dbDEPC. For all inclusive representation, circle surface areas in B and C are disproportional so that small numbers can also be shown in the graph.
We compared the content of dbDEPC with that of two existing cancer-associated gene databases (CGDCP, CGC). Most differentially expressed genes in dbDEPC are newly identified, with only 11% of genes shared with these two external databases (Figure 1B). For 106 genes validated by LTP experiments, only 8% overlap with external databases (Figure 1B). These results indicate that dbDEPC will be an important complement to previous databases.
We also explored whether differentially expressed genes in dbDEPC were associated with genomic variations. As shown in Figure 1C, 99% of dbDEPC genes have known SNPs (dbSNP database) and 41% of genes have known structural variations in the human genome (including copy number variations, inversions and insert-deletions; DGV database). This rate of DNA-level variation is significantly higher than that seen for general human genes, by Fisher's; exact test (P < 0.0001). This result may offer a starting point for explaining the mechanism of protein expression change in cancers.
DATABASE UTILITY
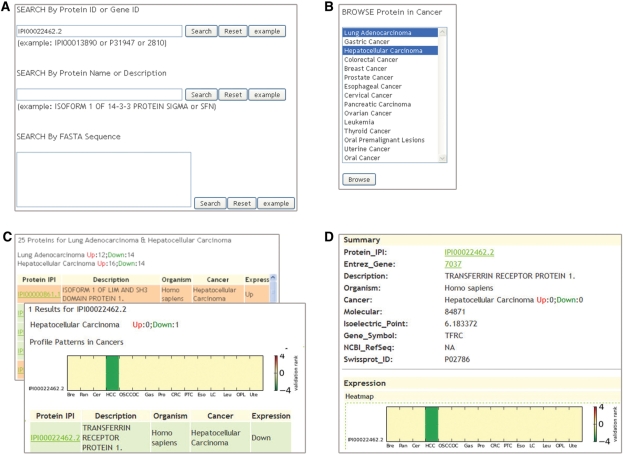
dbDEPC provides two types of searching models on its search page (Figure 2A). One is text search, supporting protein IPI identifier, Entrez gene ID or protein description. Another is sequence similarity search by BLASTP. dbDEPC’s browsing function is implemented via the browse page (Figure 2B). Users can select one or multiple cancers to browse differentially expressed proteins. Search and browse results list basic information for protein matches (Figure 2C), such as IPI ID, description, related cancer and expression change (up or down). More detailed information for each protein can be accessed by clicking the protein ID link, as shown in Figure 2D (and detailed in Supplementary Figure S1). Detailed information contains three sections: summary, expression and annotation (Supplementary Figure S1). The summary section summarizes expression changes seen in different cancers, as well as providing protein description, IDs, molecular weight and isoelectric point. The expression section plots a heatmap to visualize the expression change of proteins in multiple cancers, and provides information about all MS experiments that have identified the protein, including the PMIDs of associated publications, cancer type, expression change (up or down), fold change, MS equipments and so on. If available, validation information (LTP, CGDCP, CGC) also is provided in the expression section. The annotation section contains diverse annotations: SNPs, genomic structure variations, domain organization, Gene Ontology, KEGG pathways and protein sequence. Taking TFRC (IPI00022462.2) as an example (shown in Supplementary Figure S1), the summary section briefly describes that TFRC is down-regulated in HCC; the expression section shows that its fold change is 0.236766 or 0.3056256 based on an LTQ-FT MS experiment from an paper with the PMID 1864787, its gene is reported as a cancer-related gene based on the CGDCP database and it has somatic mutations in non-Hodgkin’s lymphoma based on the CGC database; and the annotation section notes 15 SNPs, copy number variations and inversion in the gene region, and provides other functional terms.
Figure 2.
The web interface of dbDEPC. (A) Search page. Proteins can be searched by IPI ID, Entrez gene ID, protein description or sequence. (B) Browse page. Users can browse proteins in multiple selected cancers. (C) Result page for search or browse function. Results summarize protein matches and provide a list of basic protein information. (D) A part of an example protein record. The whole protein page can be found in Supplementary Figure S1, consisting of summary, expression, and annotation sections.
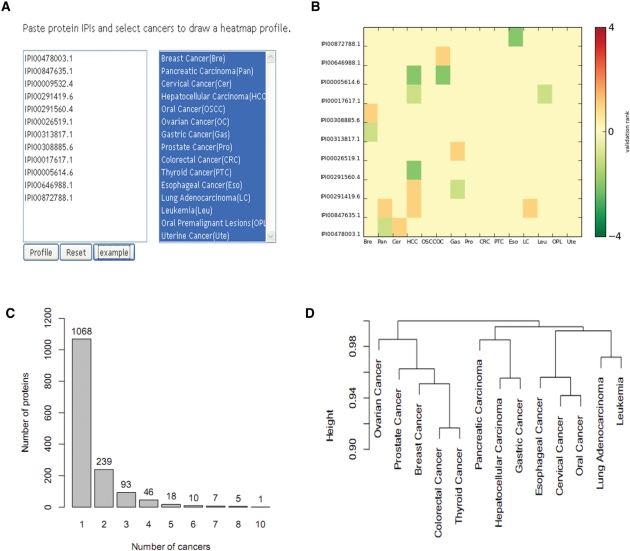
Finally, dbDEPC’s profile page can assist users to investigate differences in protein profile among different cancers. If users input a protein list and select cancers of interest on the profile page (Figure 3A) the database will return a heatmap visualizing the expression change of proteins in multiple cancers (Figure 3B) and a table listing all differentially expressed proteins. Up- and down-regulation are represented by red and green color, respectively. The color grade is dependent on a ‘validation rank’, which is calculated based on number of data sources (MS, LTP, CGDCP, CGC). Deeper color denotes more sources that relate the protein with a particular cancer.
Figure 3.
(A) Profile page. Users can input a protein list and select cancers of interest to produce an expression heatmap. (B) Example heatmap of differentially expressed proteins in multiple cancers. (C) Number of proteins in multiple cancers. (D) Clustering of cancers based on differentially expressed protein profiles.
DATABASE AVAILABILITY
All data in dbDEPC are freely available for download without password protection for academic users. dbDEPC can be accessed via http://dbdepc.biosino.org/index/, and data can be downloaded through the download page as text files.
DISCUSSION AND CONCLUSION
Differentially expressed proteins in cancers are often regarded as candidate biomarkers for diagnosis, prognosis and prediction of response to therapy. Data in dbDEPC (Figure 3C) show that some proteins have differential expression in only one cancer (specific proteins), while other proteins have differential expression in multiple cancers (common proteins). For example, the HSPA5 protein (78 kDa glucose-regulated protein, IPI00003362.2) is up-regulated in seven cancers, down-regulated in two cancers and shows conflicting experimental results in another cancer. Specific proteins may be candidate diagnostic biomarkers for specific cancers, whereas common proteins could be regarded as biomarkers for a group of cancers. dbDEPC increases the reliability of information on cancer biomarkers by curating proteomics data from multiple experiments. In addition, dbDEPC is the first database to emphasize protein-level expression change in cancers based on high-throughput MSy experiments.
In dbDEPC, the integration of data from multiple cancers also allows for comparison among cancers based on protein expression patterns. Using the expression profiles of 1487 human proteins in 13 cancers (disregarding two cancers with <50 differentially expressed proteins), we obtained a hierarchical clustering by Jaccard similarity coefficient (Figure 3D). The associations among some cancers might suggest underlying functional interaction. For example, breast cancer clusters with ovarian cancer and prostate cancer. This may imply that sex-hormone-related cancers share some common biomarkers. Data resources such as dbDEPC may facilitate development of a more systemic understanding of the relationships among different cancers at the protein expression level.
Moreover, each protein in dbDEPC is annotated with any known variations in its gene, allowing for exploration of possible underlying mechanism for protein expression changes in cancer. In fact, 99% of the genes collected in dbDEPC have known SNPs and 41% have known structural variations in the human genome (including copy number variations, inversions and insert-deletions). Additionally, most differentially expressed genes in dbDEPC are newly identified, with only 11% of genes overlapping with two external databases. In conclusion, dbDEPC is aimed to become an important reference database for differentially expressed proteins in human cancers. This new resource may facilitate cancer research and contribute to biomarker discovery.
FUTURE DEVELOPMENT
With more and more cancer proteomics experiments being performed, we plan to update dbDEPC annually. The current dbDEPC collects primarily human proteins. Model organisms are much easier to study and disease models have contributed significantly to cancer research (24). Thus, we also plan to add data from mouse and rat. Previously we have established a cancer-related gene expression signature database (under submission). Another important extension in the future will be to explore the association between RNA levels as recorded in the previous database and protein levels in the present database.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National High-Tech R&D Program (2009AA02Z304, 2007AA02Z304); National Basic Research Program (2010CB912702, 2009CB918404, 2006CB910700); Key Research Program (CAS) (KSCX2-YW-R-112); China National Key Projects for Infectious Disease (2008ZX10002-021); National Natural Science Foundation of China (30900272); Shanghai Natural Science Foundation (08ZR1415800); Special Start-up Fund for CAS President Award Winner (to G.D.). Funding for open access charge: National High-Tech R&D Program (2009AA02Z304).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
The authors acknowledge Lynne Berry from the Vanderbilt Cancer Biostatistics Center for her editing work.
REFERENCES
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 3.Kato K, Yamashita R, Matoba R, Monden M, Noguchi S, Takagi T, Nakai K. Cancer gene expression database (CGED): a database for gene expression profiling with accompanying clinical information of human cancer tissues. Nucleic Acids Res. 2005;33:D533–536. doi: 10.1093/nar/gki117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elfilali A, Lair S, Verbeke C, La Rosa P, Radvanyi F, Barillot E. ITTACA: a new database for integrated tumor transcriptome array and clinical data analysis. Nucleic Acids Res. 2006;34:D613–616. doi: 10.1093/nar/gkj022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr. Protoc. Hum. Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10, Unit 10 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semmes OJ, Malik G, Ward M. Application of mass spectrometry to the discovery of biomarkers for detection of prostate cancer. J. Cell Biochem. 2006;98:496–503. doi: 10.1002/jcb.20855. [DOI] [PubMed] [Google Scholar]
- 7.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 8.Pan J, Chen HQ, Sun YH, Zhang JH, Luo XY. Comparative proteomic analysis of non-small-cell lung cancer and normal controls using serum label-free quantitative shotgun technology. Lung. 2008;186:255–261. doi: 10.1007/s00408-008-9093-7. [DOI] [PubMed] [Google Scholar]
- 9.Ueda K, Katagiri T, Shimada T, Irie S, Sato TA, Nakamura Y, Daigo Y. Comparative profiling of serum glycoproteome by sequential purification of glycoproteins and 2-nitrobenzensulfenyl (NBS) stable isotope labeling: a new approach for the novel biomarker discovery for cancer. J. Proteome Res. 2007;6:3475–3483. doi: 10.1021/pr070103h. [DOI] [PubMed] [Google Scholar]
- 10.Chen G, Li A, Zhao M, Gao Y, Zhou T, Xu Y, Du Z, Zhang X, Yu X. Proteomic analysis identifies protein targets responsible for depsipeptide sensitivity in tumor cells. J. Proteome Res. 2008;7:2733–2742. doi: 10.1021/pr7008753. [DOI] [PubMed] [Google Scholar]
- 11.Bouchal P, Roumeliotis T, Hrstka R, Nenutil R, Vojtesek B, Garbis SD. Biomarker discovery in low-grade breast cancer using isobaric stable isotope tags and two-dimensional liquid chromatography-tandem mass spectrometry (iTRAQ-2DLC-MS/MS) based quantitative proteomic analysis. J. Proteome Res. 2009;8:362–373. doi: 10.1021/pr800622b. [DOI] [PubMed] [Google Scholar]
- 12.Deng SS, Xing TY, Zhou HY, Xiong RH, Lu YG, Wen B, Liu SQ, Yang HJ. Comparative proteome analysis of breast cancer and adjacent normal breast tissues in human. Gen. Proteom. Bioinfo. 2006;4:165–172. doi: 10.1016/S1672-0229(06)60029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho J, Kong JW, Choong LY, Loh MC, Toy W, Chong PK, Wong CH, Wong CY, Shah N, Lim YP. Novel breast cancer metastasis-associated proteins. J. Proteome Res. 2009;8:583–594. doi: 10.1021/pr8007368. [DOI] [PubMed] [Google Scholar]
- 14.Yocum AK, Busch CM, Felix CA, Blair IA. Proteomics-based strategy to identify biomarkers and pharmacological targets in leukemias with t(4;11) translocations. J. Proteome Res. 2006;5:2743–2753. doi: 10.1021/pr060235v. [DOI] [PubMed] [Google Scholar]
- 15.Ralhan R, Desouza LV, Matta A, Chandra Tripathi S, Ghanny S, Dattagupta S, Thakar A, Chauhan SS, Siu KW. iTRAQ-multidimensional liquid chromatography and tandem mass spectrometry-based identification of potential biomarkers of oral epithelial dysplasia and novel networks between inflammation and premalignancy. J. Proteome Res. 2009;8:300–309. doi: 10.1021/pr800501j. [DOI] [PubMed] [Google Scholar]
- 16.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 17.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Ding G, Xie L, Li Y. PAnnBuilder: an R package for assembling proteomic annotation data. Bioinformatics. 2009;25:1094–1095. doi: 10.1093/bioinformatics/btp100. [DOI] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet. Genome Res. 2006;115:205–214. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
- 24.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat. Rev. Cancer. 2007;7:645–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]