Abstract
EuPathDB (http://EuPathDB.org; formerly ApiDB) is an integrated database covering the eukaryotic pathogens of the genera Cryptosporidium, Giardia, Leishmania, Neospora, Plasmodium, Toxoplasma, Trichomonas and Trypanosoma. While each of these groups is supported by a taxon-specific database built upon the same infrastructure, the EuPathDB portal offers an entry point to all these resources, and the opportunity to leverage orthology for searches across genera. The most recent release of EuPathDB includes updates and changes affecting data content, infrastructure and the user interface, improving data access and enhancing the user experience. EuPathDB currently supports more than 80 searches and the recently-implemented ‘search strategy’ system enables users to construct complex multi-step searches via a graphical interface. Search results are dynamically displayed as the strategy is constructed or modified, and can be downloaded, saved, revised, or shared with other database users.
INTRODUCTION
The Eukaryotic Pathogen Database (EuPathDB) has been developed as a Bioinformatics Resource Center (BRC), with support from the US National Institutes of Health (NIH). Additional funding for a kinetoplastid component (TriTrypDB) was recently provided by the Bill and Melinda Gates Foundation, in coordination with independent support for kinetoplastid annotation from the Wellcome Trust. EuPathDB has undergone a dramatic expansion over the past two years, evolving from an apicomplexan-specific database (ApiDB) (1) to encompass additional non-apicomplexan pathogens of human and veterinary importance, most of which are classified as category B biodefense pathogens by the National Institute of Allergies and Infectious Diseases (NIAID). The eukaryotic pathogens included in EuPathDB are a major source of morbidity and mortality worldwide, causing millions of deaths and an immense economic burden.
The current version of EuPathDB (version 2.1) serves as a portal for GiardiaDB (2), CryptoDB (3), PlasmoDB (4), ToxoDB (5), TrichDB (2) and TriTrypDB (companion paper in this issue). These databases provide access to genomic and functional genomic data for 22 species (Table 1). In addition, EuPathDB provides access to genomic and/or expressed sequence tag data for additional related apicomplexan and trypanosomatid species (Table 1). Users can access functional data directly via the component web sites or through EuPathDB where query execution retrieves data from the component sites via Web services. One advantage of running searches in EuPathDB is the ability to leverage functional data from the diverse organisms we support and combine the search results based on orthology (6). The diversity of functional genomics data available through EuPathDB continues to expand dramatically (Table 1) making it essential to provide clients with an interface that is functional, user-friendly and sophisticated (Figure 1). To this end, we have recently introduced a novel search system enabling users to build complex queries comprised of individual searches (steps) that are linked with each other using set operations (union, intersection, and minus) (Figure 1D).
Table 1.
Species and data types included in the EuPathDB family of databases
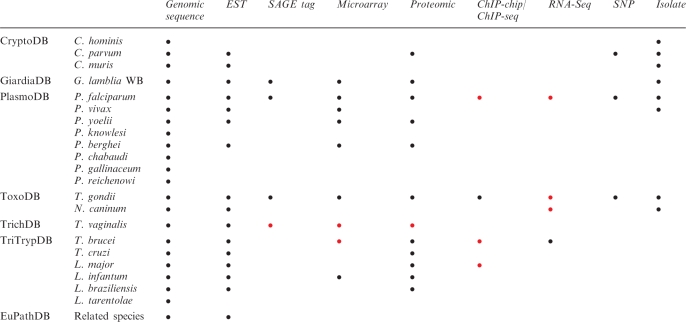 |
Data anticipated by March, 2010 is indicated in red. EuPathDB related species include genomic sequences for Theileria parva and annulata and EST sequences for Eimeria spp., Babesia spp., Sarcocystis spp., Theileria spp., Neospora hughesi and Gregarina niphandordes.
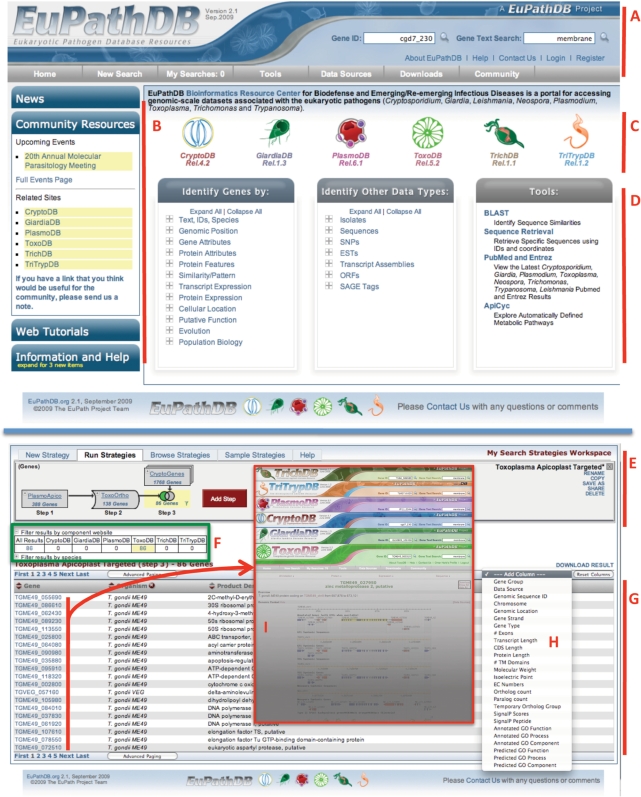
Figure 1.
Screen shots from the EuPathDB database. (A) Interactive banner present on all EuPathDB web pages, including quick search windows and a tool bar (grey). (B) Side bar components contain expandable sections for release notes, community resources, tutorials and help (new items are highlighted with a yellow alert). (C) Hyperlinked logos of the EuPathDB component sites direct users to component databases. (D) Gene searches, non-gene entity searches, such as ESTs, ORFs of genome sequence and useful tools (BLAST against genomes in EuPathDB, PubMed records pertaining to EuPathDB organisms etc.). Note: clicking on the “+” symbol reveals a list of searches available within each category. (E) A multi-step search strategy, leveraging orthology to identify apicoplast targeted genes in Toxoplasma. (F) Summary table that allows filtering results based on component sites. (G) The results of a search strategy appear dynamically as the strategy is built and modified. (H) Additional columns of data can be added to results list. (I) Individual gene pages in the component sites can be accessed by clicking on their gene IDs.
WHAT IS NEW IN EuPathDB
Over the past two years EuPathDB (formerly ApiDB) has evolved from an apicomplexan-specific resource (CryptoDB, PlasmoDB and ToxoDB) to encompass protozoan pathogens outside this phylum, doubling its repertoire of functional genomic databases (GiardiaDB and TrichDB in early 2008, and TriTrypDB in early 2009). Concurrent with this expansion, EuPathDB has undergone dramatic changes specifically designed to enhance data accessibility and provide database users with a graphical query-building interface that effectively creates a venue for constructing complex search strategies with relative ease.
The EuPathDB homepage provides the user with quick and easy access to information by providing specific, functional sections (Figure 1A–D), a design feature that is replicated across EuPathDB component sites. The top part of the page (banner) remains constant throughout EuPathDB Web pages, providing quick access to text- and gene ID-based searches, links to useful pages (help and information pages, a ‘Contact Us’ link, registration/login), and a tool bar (gray) with links to access diverse searches (see below), the user’s personal search history, tools, downloads, data sources, and other links (Figure 1A). The left side of the home page (Figure 1B) provides a series of expandable windows containing news items (release notes, etc.), tutorials (demonstrating the website), community resources, and additional information and help. The number of new items added since the user’s last visit is indicated by yellow numbers. The middle of the page includes hyperlinked logos for all EuPathDB component sites, providing direct access to taxon-specific databases (Figure 1C). Below, three sections provide access to queries and tools (Figure 1D): (i) gene-centric searches allowing the identification of genes based on text terms, species, transcript and protein expression, GO terms, EC numbers, user defined protein motif patterns, phylogenetic profile, etc.; (ii) other data type searches such as isolates and their geographic or clinical origin, expressed sequence tags (ESTs), single nucleotide polymorphism (SNP) data, etc.; (iii) access to useful tools such as BLAST, the sequence retrieval tool, metabolic pathways and PubMed records.
Unlike queries available through the organism-specific component databases, where users are restricted to datasets available in that particular database, the EuPathDB user has the opportunity to explore data from all organisms represented on any component site. This expanded range provides the ability to transform a set of results from one organism into its orthologs in another, making it possible to generate comparable lists of results in organisms where particular functional data are lacking. Execution of queries in EuPathDB starts by selecting a type of search from the options shown in Figure 1D. A strategy expands upon the first search (Step 1) by adding additional searches using set operators such as union, intersect and minus (graphically displayed with Venn diagrams, Figure 1E). Any step in the strategy can be transformed into orthologs from selected species of interest. Figure 1E presents an example in which orthology is leveraged between organisms to generate a list of potential apicoplast (plastid-like organelle) targeted genes in Toxoplasma using information from other species. In step one, genes predicted to target to the parasite-specific apicoplast arganelle in P. falciparum are obtained based on available targeting information data (7). This list is transformed into orthologs in the related species T. gondii in step two, yielding 138 candidates. In step three, this list is restricted by subtracting genes found in Cryptosporidium (a related organism that has lost its apicoplast), leaving 86 possible Toxoplasma apicoplast-targeted genes. Many of these genes have been experimentally proven to target to the apicoplast, validating this approach. Analogous search strategies can be used to generate a wide variety of potentially useful gene lists, such as: genes in Toxoplasma and Giardia that are orthologs of oocyst genes in Cryptosporidium and Plasmodium (for which functional data are available); or orthologous genes expressed in the Plasmodium and Trypanosoma arthropod stages; or genes for which reagents are available (such as antibodies) in other organisms.
Steps in a strategy can be renamed, revised, viewed, deleted, downloaded or expanded into sub-strategies. In addition, whole strategies can be renamed, saved, copied, closed/opened, deleted or shared with others via a uniquely generated URL (a recipient of the URL can click on it to generate the strategy in their own workspace). Previously executed strategies can be accessed via ‘Browse Strategies’ by clicking on the appropriate tab (Figure 1E).
Results are dynamically updated below the strategy in an interactive summary table (Figure 1F, green box) that allows a user to click on the number of genes under the component database to filter the results revealing only genes from that database or all databases by clicking on the ‘all results’ link. The actual gene list appears dynamically below the summary table (Figure 1G) and can be sorted, expanded with additional columns of data (Figure 1H) or downloaded. Detailed gene information can be viewed by visiting specific gene pages, which are accessed by clicking on the gene IDs in the result list (Red box in Figure 1I).
In an effort to promote community involvement in annotation, users can enter comments on gene and isolate record pages. Such comments can include PubMed IDs (references are automatically retrieved for final display in the comment), GenBank accession numbers, files (such as images), and comments that apply to multiple gene or isolate records can be easily replicated by adding the related IDs to the comment form. Comments immediately appear on record pages and become indexed and searchable via the text search. This feature has positively contributed to genome curation efforts in general and has specifically improved the official annotation of the P. falciparum and Trypanosomatidae genomes (see companion TriTrypDB paper for further information). Users of EuPathDB and its family of databases can also upload files to a community file repository that provides a venue for users to share data that are not linked to a specific record, such as, protocols.
FUTURE DIRECTIONS
The EuPathDB family of databases is expected to expand further over the coming years, incorporating new species including the Amoebozoa (Entamoeba, Acanthamoeba), Microsporidia, and additional Apicomplexans, Kinetoplastida and Diplomonads. Features and tools will continue to be introduced and improved, such as the ability to make connections between diverse data types, a basket for users to place cherry-picked genes for subsequent manipulation and weighting of queries to enable more effective analysis of complex strategy results. Of course, we also anticipate that data will continue to be deposited at an ever-increasing rate, including additional genome sequences, microarray data, probe-based hybridization and sequencing data (e.g. ChIP-chip and RNA-Seq), proteomics data, isolate data, phenotype information, metabolomic data, etc.
FUNDING
EuPathDB is funded with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN266200400037C (to D.S.R., C.J.S. and J.C.K.). The TriTrypDB component is supported by grant no. 50096 from the Bill and Melinda Gates Foundation. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to acknowledge the contribution of numerous members of the eukaryotic pathogen research community, in the form of advice, suggestions and/or data—often made available to the community via EuPathDB and its components in advance of publication.
REFERENCES
- 1.Aurrecoechea C, Heiges M, Wang H, Wang Z, Fischer S, Rhodes P, Miller J, Kraemer E, Stoeckert CJ, Jr, Roos DS, et al. ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 2007;35:D427–D430. doi: 10.1093/nar/gkl880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aurrecoechea C, Brestelli J, Brunk BP, Carlton JM, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, et al. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 2009;37:D526–D530. doi: 10.1093/nar/gkn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heiges M, Wang H, Robinson E, Aurrecoechea C, Gao X, Kaluskar N, Rhodes P, Wang S, He CZ, Su Y, et al. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 2006;34:D419–D422. doi: 10.1093/nar/gkj078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foth BJ, Ralph SA, Tonkin CJ, Struck NS, Fraunholz M, Roos DS, Cowman AF, McFadden GI. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003;299:705–708. doi: 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]