Abstract
The Comprehensive Microbial Resource or CMR (http://cmr.jcvi.org) provides a web-based central resource for the display, search and analysis of the sequence and annotation for complete and publicly available bacterial and archaeal genomes. In addition to displaying the original annotation from GenBank, the CMR makes available secondary automated structural and functional annotation across all genomes to provide consistent data types necessary for effective mining of genomic data. Precomputed homology searches are stored to allow meaningful genome comparisons. The CMR supplies users with over 50 different tools to utilize the sequence and annotation data across one or more of the 571 currently available genomes. At the gene level users can view the gene annotation and underlying evidence. Genome level information includes whole genome graphical displays, biochemical pathway maps and genome summary data. Comparative tools display analysis between genomes with homology and genome alignment tools, and searches across the accessions, annotation, and evidence assigned to all genes/genomes are available. The data and tools on the CMR aid genomic research and analysis, and the CMR is included in over 200 scientific publications. The code underlying the CMR website and the CMR database are freely available for download with no license restrictions.
INTRODUCTION
The large volumes of genomic data being produced with more efficient and cost effective new sequencing technologies will increase the rate of scientific discovery only if investigators are able to find the data that is of specific interest to their research. Today at GenBank (1) (http://www.ncbi.nlm.nih.gov/Genbank/), the central repository of genome data, there are over 900 complete, publicly available bacterial and archaeal genomes from hundreds of sequencing centers. The computations and searches across multiple genomes that allow scientists to effectively mine this genomic data are only possible when annotation is uniformly applied to all bacterial sequences. Since 2000, the J. Craig Venter Institute [JCVI; JCVI will be used throughout to denote The J. Craig Venter Institute or its predecessor organizations, including The Institute for Genomic Research (TIGR) that was merged with JCVI in 2006] has provided the Comprehensive Microbial Resource or CMR (http://cmr.jcvi.org). The CMR is a central repository containing the sequence and original annotation of complete prokaryotic genomes as well as standard automated annotation across all genomes and precomputed homology searches to allow meaningful genome comparisons. The CMR currently contains 571 genomes and over 50 tools are available to utilize and mine this genomic data. Some of these genomes have websites provided by their sequencing center, and all are available at GenBank. However, comparative genomics and searches across more than a single genome are enhanced when genomic data is located in a common repository with consistent annotation and bioinformatics tools that serve the diverse needs of the scientific community. The CMR has provided such a resource to the prokaryotic scientific community for over nine years.
PROKARYOTIC ANNOTATION DATA ISSUES AND RESOLUTION
Sequencing centers employ a variety of gene finding methods and data management strategies. Similarly, there is considerable variation among annotation groups regarding how function, gene symbols, Enzyme Commission (EC) numbers (2), Gene Ontology (GO) terms (3) and functional role category assignments are produced. Variable approaches to annotation are unavoidable given the size and diversity of microbial genomics; however, robust comparisons across all prokaryotic genomes are facilitated when annotation is systematically assigned by applying identical data types. An additional challenge to robust genome comparison are the inconsistencies (4) found across the annotation files submitted to GenBank.
To create consistent data in the CMR, a two-stage approach is employed. The first stage is to carefully extract data from the GenBank records of complete genomes and categorize them based on rigorous data types. Data types such as EC numbers can be located in different tagged fields in the records for different genomes; a customized text parser is used to capture critical elements such as genes, gene product and EC numbers, and load them into explicit fields of a relational database. The second stage is to assign additional, consistent annotation across all genomes using an automated pipeline. The automated annotation pipeline assigns function, gene symbols, JCVI functional role categories [based on Monica Riley’s Escherichia coli functional classification system (5)], EC numbers, GO terms and related evidence in a consistent manner, allowing for reliable comparisons across uniform annotation for all CMR genomes.
Annotation from the original sequencing center extracted from GenBank is defined as the ‘primary’ annotation for each genome. The primary annotation and the annotation from the JCVI automated pipeline (called secondary or JCVI annotation) are stored separately in the database, and genes agreed on by both the primary and secondary annotation are linked. Nearly all of the CMR tools can be based on either the primary or the automated annotation, with the option to toggle between the two types. However, because the secondary annotation is automatically generated, the primary annotation is preferentially displayed throughout the CMR.
AUTOMATED ANNOTATION FOR CMR GENOMES
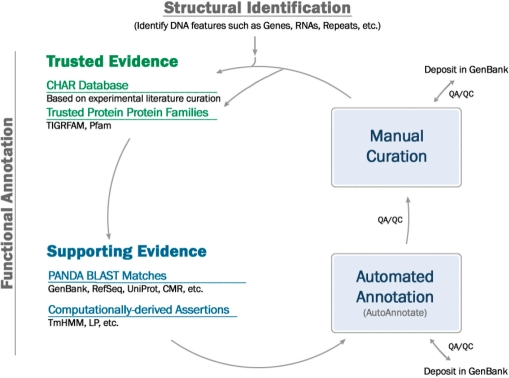
To create the secondary or JCVI annotation for all CMR genomes, JCVI employs an automated annotation pipeline that identifies genome features in the raw DNA sequence, gathers evidence for function of the features, and assigns functional annotation based on the weight of the evidence. Annotation is an ongoing cyclical process and annotation of new genomes or re-annotation of older genomes is improved as new trusted evidence is produced. Figure 1 shows an overview of the JCVI annotation process.
Figure 1.
JCVI Prokaryotic Genome Annotation. The annotation pipeline begins with DNA feature identification and then goes into the cyclical process of functional annotation. Automated annotation is based on trusted and supporting evidence. Manual curation of proteins or, more often today, of trusted evidence types further strengthens the annotation pipeline and can be used when annotation is regenerated. Genomes in the CMR not originally sequenced at JCVI are deposited into the CMR after automated annotation.
DNA feature identification
Glimmer3 (6) is used to predict protein coding sequences (CDS), tRNAs are identified with the tRNAscan tool (7), rRNA genes and other structural RNAs are identified directly from BLAST (8) matches to Rfam (9), a database of non-coding RNA families.
Evidence for functional annotation
JCVI uses a combination of trusted evidence types which provide consistent functional annotation and can be transferred onto genes with high confidence in an automated fashion. The two major trusted evidence types used in the annotation pipeline are:
CHAR database: JCVI’s CHAR is a curated database of experimentally verified proteins, source publications, and functional annotations. Each protein entry has detailed annotation including function, gene symbol, and GO terms and evidence codes.
Trusted protein families: these families currently include JCVI’s TIGRFAM protein family models (10) and Pfams (11), both built on Hidden Markov Models (HMMs). JCVI is collaborating with other centers to consolidate, validate and incorporate similar high quality protein classification systems [e.g. NCBI’s PRK clusters (12)].
Supporting evidence for the annotation pipeline includes:
BLAST searches against PANDA: PANDA is JCVI’s internal repository of non-redundant and non-identical protein and nucleotide data pulled from public databases that include the latest assembly and protein sequences (e.g. GenBank, RefSeq, UniProt, Protein Data Bank, CMR). PANDA is available on the JCVI FTP site (ftp://ftp.jcvi.org/pub/data/panda).
Computationally derived assertions: computations integral to the pipeline include derived physical and chemical metrics including lipoprotein signals (LP) and transmembrane helices [TmHMM, (13)].
AutoAnnotate
AutoAnnotate weighs the evidence from a precedence-ordered list of evidence types—the CHAR database, trusted protein families, best protein BLAST matches from PANDA and computationally derived assertions—to annotate each protein by assigning, where possible, a function, gene symbol, EC numbers, JCVI functional role category and GO terms. AutoAnnotate and the databases on which AutoAnnotate runs are freely available for download and installation via the open source repository SourceForge (https://sourceforge.net/projects/prokfunautoanno/).
THE CMR WEB RESOURCE
The CMR comparative database contains complete, public bacterial and archaeal genomes, with a web interface that allows for a wide variety of data retrievals pertaining to inter- and intragenomic relationships for comparative genomics, genome diversity and evolutionary studies. Retrievals can be based on a number of different properties, including molecular weight, hydrophobicity, GC-content, functional role assignments and taxonomy. The CMR interface is designed to make it easy for users to create complex database queries using menu-driven web pages. The CMR has special web-based tools to allow analysis using pre-computed homology searches (i.e. All versus All searches generated using BLASTP), whole genome dot-plots, batch downloading and searches across genomes using a variety of data types.
CMR data model
The Omniome is the production database underlying the CMR, and it holds all of the annotation for each of the CMR genomes, including DNA sequences, proteins, RNA genes and many other features. Associated with each of these DNA features in the Omniome are the coordinates, nucleotide and protein sequences (where appropriate), and the DNA molecule and organism with which the feature is associated. Also available are evidence types associated with annotation such as HMMs (TIGRFAMs and Pfams), BLAST, InterPro (14), NCBI COG (15) and PROSITE (16), individual gene attributes and identifiers from other centers such as GenBank and Swiss-Prot/UniProt (17), manually curated information on each genome and the precomputed All versus All searches.
The CMR tools
New users can learn about the functionality and navigation of the CMR website by utilizing the CMR user manual, Frequently Asked Questions and an on-line tutorial, all available off of the CMR home page. In addition, each page on the CMR provides detailed descriptions on the content and usage of the tool under an information icon ‘i’. Over 50 different tools are available on the CMR, broken into seven major categories:
Searches: Searches allow users to find genes, genomes, sequences, or text from data stored in the CMR. Searches to find genes based on annotation (i.e. locus identifier, functional name, gene symbol), alternate accessions (i.e. GenBank, SwissProt), and evidence or role categories (i.e. EC numbers, GO terms, TIGRFAMs, Pfams, Interpro, Prosite, NCBI COGs and JCVI functional role categories) across all genomes in the CMR are available, as are BLAST searches against the CMR database, HMM sequence searches against TIGRFAMs and Pfams, protein motif searches, and the ability to retrieve nucleotide sequence or list of genes between two coordinates.
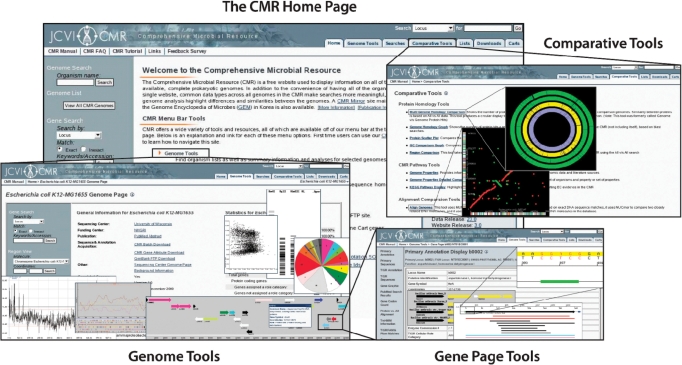
Genome tools: Genome level information (Figure 2, Genome Tools) includes graphical displays showing genes placed linearly on regions of the chromosome, or as a complete circle. Other pages give overviews of pathways and subsystems utilizing JCVI’s Genome Properties database (18) and KEGG biochemical pathway maps (19). Codon usage tables, GC plots, computer generated 2D gels, restriction digest tools, JCVI functional role category graphs, and tables summarizing information such as average gene size or numbers of coding regions are also available.
Figure 2.
The CMR. The CMR provides both prokaryotic genome data and analytic tools. Examples from three of the major groups of tools available are shown: Genome, Gene Page and Comparative Tools.
Comparative tools: Comparative tools include homolog analyses and genome alignment displays (Figure 2, Comparative Tools). For example, a whole genome alignment between two bacteria using MUMmer (20) can be seen in a dotplot showing all the genes that are homologous between any two genomes the user selects; schematically, one can see large-scale conserved synteny as well as inversions and translocations in this display. Other displays show protein homology across genomes on a whole genome scale, or focus on a particular region of the genome. The Multi-Genome Homology tool calculates and displays homologous proteins across a single reference genome and up to 15 comparison genomes, with a summary of all proteins unique to the reference and in common with the comparison genomes. The Region Comparison tool aligns the overall best matching regions from other genomes to a user selected reference region.
Lists: The three main categories of lists available are gene lists (e.g. all genes by JCVI functional role category), lists of other genomic elements (e.g. all RNAs in a genome, all intergenic regions in a genome) and evidence lists (e.g. lists of all EC numbers, TIGRFAMs, Pfams and NCBI COGs in the CMR).
Downloads: Both Batch Download and the Gene Attribute Download tools are available. The Batch Download allows users to get a FASTA file of the nucleotide or protein sequence for a set of genes. The Gene Attribute Download allows users to download over 20 different gene attributes (e.g. coordinates, gene symbol, product name, EC number, GenBank ID) for a set of genes. To select genes for these tools the user can upload a list of accessions or select all genes from an organism and/or JCVI functional role category.
Carts: Two carts are currently available, a Genome Cart and a Gene Cart. The Genome Cart allows users to choose their genomes of interest; these genomes are then preselected when a user comes to an organism selection menu. The Gene Cart allows users to select genes of interest while perusing the CMR and for use in later retrievals. Users can select genes into the Gene Cart from any CMR page that shows a list of genes; selected genes can be viewed and downloaded using the Batch Download and Gene Attribute Download tools from the Gene Cart page.
Gene pages: At the gene level (Figure 2, Gene Page Tools), users are able to view the annotation given to the gene both from the primary and automatic annotation including the product name, gene symbol, EC number, GO terms, JCVI functional role category assignment, DNA sequence and protein sequence. Users may view the TmHMM profile, links to other resources such as UniProt and GenBank, secondary structure, third position GC-Skew and many other displays.
CMR use cases
A review of articles referencing the CMR indicates a variety of use cases. Researchers retrieve gene sequences (21–23), download whole genomes (24) and BLAST against the CMR sequence databases (25). Many scientists use the CMR to provide JVCI functional role category classification across one or more organisms (26–30), showing the importance of having the standard functional classification across all genomes that the CMR provides. The CMR is used for functional classification of genes on microarrays (29) and for microarray design (31). Identifying codon usage and tRNA gene copy number (32), operon identification to provide a genomic link between genes to support laboratory results (33), and the identification of novel genes not called in the original GenBank annotation (34) are other ways the CMR is aiding researchers.
Scientists are using the CMR for comparative genomics including the analysis of potential intragenome transfers with the Multi Genome Homology tool (35), analysis of flanking DNA using the Region View tool (36) and identification of proteins in other bacteria that are similar to a test set using the Genomes Region Comparison tool (37).
CMR updates
New genomes are added to the CMR two to four times per calendar year. Genomes released at GenBank since the last update are downloaded and put through automated annotation and added to the CMR database, the Omniome. JCVI genomes published or released to GenBank since the last update are added to the Omniome, and All versus All homology searches are performed on all genomes. These precomputed BLASTP searches are used throughout the CMR for comparative analysis. Once all data in the Omniome is validated by a series of consistency checks it is tagged and released to the CMR website as a versioned data update. New releases are advertised on the CMR home page.
The CMR currently contains 571 organisms, while GenBank has more than 900 complete prokaryotic genomes. JCVI is currently working on updating the number of genomes in the CMR to reflect and keep up with the complete genomes at GenBank. JCVI expects the CMR to contain over 800 genomes by early 2010, and be caught up with GenBank by mid 2010.
CMR 3-tier system
The CMR is implemented in a ‘3-tier’ architecture written in Perl. The tiers of this architecture are the presentation tier (i.e. user interface), the functional process logic tier, and the data storage and access tier (i.e. database tier). This architecture is ideal for re-use into other applications; developers can take advantage of existing functions easily and complex retrievals from the database are simple. Overall this system provides an environment where developers can add functions quickly while providing the ability to merge changes into the mature shared codebase.
CMR statistics
Over the past year, an average of 16 000 unique users per month have accessed the CMR, the average number of visits was 29 000 per month (each user averaging 1.8 monthly visits), and the average number of page hits was 270 000 per month (each user averaging 16 CMR page hits per month). The majority of traceable CMR users come from US educational (.edu) domains; in total, people from over 100 countries use the website on a monthly basis. A survey of scientific publications from the past nine years shows over 200 publications report using the CMR (e.g., 38–47).
AVAILABILITY OF THE CMR DATABASE
Under the CMR Downloads menu are freely available, restriction free copies of all of the CMR Perl web applications, as well as the Omniome CMR database, either as a MySQL database or in tab delimited files. A schema is available showing the database table relationships and detailed descriptions of the tables and rows. Using the downloadable database and Perl programs, local installations of the CMR are possible and two CMR mirror sites, one public and one private, have been set up by two centers. The CMR database is routinely downloaded to provide the major genome data feed for the BioCyc (48) collection of Pathway/Genome Databases.
For users who do not wish to download the whole underlying database, the CMR provides all tables displayed throughout the website with a ‘Download’ button allowing the user to open the table as a tab delimited file, exportable to a spreadsheet program. In addition, all graphics on the CMR have links to the underlying data in downloadable table format.
PROKARYOTIC ANNOTATION AND ANALYSIS COURSE
Since 2002, JCVI has offered community training in annotation of prokaryotic genomes (http://www.jcvi.org/AnnotationClass/) four times a year. The course starts with an extensive overview of the prokaryotic annotation process at JCVI including gene finding, similarity searching, evidence interpretation, protein naming, and the GO system. Attendees are given a detailed tutorial on JCVI’s manual annotation tool Manatee and guided through the manual annotation of several genes. Finally, attendees receive in-depth look at the CMR, its features and the many analyses possible with the tools on the site. Since 2002, 242 scientists and graduate- and undergraduate students have attended.
FUNDING
Department of Energy [DE-FC02-95ER61962, DE-FG02-01ER63203]; and the National Science Foundation [DBI-0110270]. Funding for open access charge: JCVI.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank the J. Craig Venter Institute Information Technology and Bioinformatics Departments for their ongoing technical, engineering and scientific support including Michael Heaney, Dan Haft, Jeremy Selengut, Scott Durkin, Susmita Shrivastava, Lauren Brinkac, Roland Richter, Peter Rosanelli and Tom Emmel; as well as the support received from former employees of The Institute for Genomic Research including William Nelson, Sam Angiuoli and Anup Mahurkar.
REFERENCES
- 1.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webb EC. Enzyme Nomenclature. San Diego, CA: Academic Press; 1992. [Google Scholar]
- 3.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kersey P, Bower L, Morris L, Horne A, Petryszak R, Kanz C, Kanapin A, Das U, Michoud K, Phan I, et al. Integr8 and Genome Reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riley M. Functions of the gene products of Escherichia coli. Microbiol. Rev. 1993;57:862–952. doi: 10.1128/mr.57.4.862-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O'N;eill K, Resch W, Resenchuk S, et al. The National Center for Biotechnology Information's; Protein Clusters Database. Nucleic Acids Res. 2009;37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 14.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bradley P, Bork P, Bucher P, Cerutti L, et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haft DH, Selengut JD, Brinkac LM, Zafar N, White O. Genome Properties: a system for the investigation of prokaryotic genetic content for microbiology, genome annotation and comparative genomics. Bioinformatics. 2005;21:293–306. doi: 10.1093/bioinformatics/bti015. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roca AI, Almada AE, Abajian AC. ProfileGrids as a new visual representation of large multiple sequence alignments: a case study of the RecA protein family. BMC Bioinformatics. 2008;9:554. doi: 10.1186/1471-2105-9-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke TFt, Clark PL. Rare codons cluster. PLoS ONE. 2008;3:e3412. doi: 10.1371/journal.pone.0003412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humbert O, Salama NR. The Helicobacter pylori HpyAXII restriction-modification system limits exogenous DNA uptake by targeting GTAC sites but shows asymmetric conservation of the DNA methyltransferase and restriction endonuclease components. Nucleic Acids Res. 2008;36:6893–6906. doi: 10.1093/nar/gkn718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbons HS, Wolschendorf F, Abshire M, Niederweis M, Braunstein M. Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway. J. Bacteriol. 2007;189:5090–5100. doi: 10.1128/JB.00163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parks AR, Peters JE. Transposon Tn7 is widespread in diverse bacteria and forms genomic islands. J. Bacteriol. 2007;189:2170–2173. doi: 10.1128/JB.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alice AF, Naka H, Crosa JH. Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus. Infect Immun. 2008;76:4019–4037. doi: 10.1128/IAI.00208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ansong C, Yoon H, Porwollik S, Mottaz-Brewer H, Petritis BO, Jaitly N, Adkins JN, McClelland M, Heffron F, Smith RD. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS ONE. 2009;4:e4809. doi: 10.1371/journal.pone.0004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durot M, Le Fevre F, de Berardinis V, Kreimeyer A, Vallenet D, Combe C, Smidtas S, Salanoubat M, Weissenbach J, Schachter V. Iterative reconstruction of a global metabolic model of Acinetobacter baylyi ADP1 using high-throughput growth phenotype and gene essentiality data. BMC Syst. Biol. 2008;2:85. doi: 10.1186/1752-0509-2-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lone AG, Deslandes V, Nash JH, Jacques M, Macinnes JI. Modulation of gene expression in Actinobacillus pleuropneumoniae exposed to bronchoalveolar fluid. PLoS ONE. 2009;4:e6139. doi: 10.1371/journal.pone.0006139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mamirova L, Popadin K, Gelfand MS. Purifying selection in mitochondria, free-living and obligate intracellular proteobacteria. BMC Evol. Biol. 2007;7:17. doi: 10.1186/1471-2148-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouillard JM, Gulari E. OligoArrayDb: pangenomic oligonucleotide microarray probe sets database. Nucleic Acids Res. 2009;37:D938–D941. doi: 10.1093/nar/gkn761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dethlefsen L, Schmidt TM. Performance of the translational apparatus varies with the ecological strategies of bacteria. J. Bacteriol. 2007;189:3237–3245. doi: 10.1128/JB.01686-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marienhagen J, Eggeling L. Metabolic function of Corynebacterium glutamicum aminotransferases AlaT and AvtA and impact on L-valine production. Appl. Environ. Microbiol. 2008;74:7457–7462. doi: 10.1128/AEM.01025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandel MJ, Stabb EV, Ruby EG. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: focus on point miscalls and artefactual expansions. BMC Genomics. 2008;9:138. doi: 10.1186/1471-2164-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slater SC, Goldman BS, Goodner B, Setubal JC, Farrand SK, Nester EW, Burr TJ, Banta L, Dickerman AW, Paulsen I, et al. Genome sequences of three agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J. Bacteriol. 2009;191:2501–2511. doi: 10.1128/JB.01779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu SW, Chen SY, Wong HC. Dynamic localization of MreB in Vibrio parahaemolyticus and in the ectopic host bacterium Escherichia coli. Appl. Environ. Microbiol. 2008;74:6739–6745. doi: 10.1128/AEM.01021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicely NI, Parsonage D, Paige C, Newton GL, Fahey RC, Leonardi R, Jackowski S, Mallett TC, Claiborne A. Structure of the type III pantothenate kinase from Bacillus anthracis at 2.0 A resolution: implications for coenzyme A-dependent redox biology. Biochemistry. 2007;46:3234–3245. doi: 10.1021/bi062299p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alice AF, Lopez CS, Lowe CA, Ledesma MA, Crosa JH. Genetic and transcriptional analysis of the siderophore malleobactin biosynthesis and transport genes in the human pathogen Burkholderia pseudomallei K96243. J. Bacteriol. 2006;188:1551–1566. doi: 10.1128/JB.188.4.1551-1566.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett CL, Palsson BO. Iterative reconstruction of transcriptional regulatory networks: an algorithmic approach. PLoS Comput. Biol. 2006;2:e52. doi: 10.1371/journal.pcbi.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc. Natl Acad. Sci. USA. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandonia JM, Kim SH. Structural proteomics of minimal organisms: conservation of protein fold usage and evolutionary implications. BMC Struct. Biol. 2006;6:7. doi: 10.1186/1472-6807-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ducey TF, Carson MB, Orvis J, Stintzi AP, Dyer DW. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J. Bacteriol. 2005;187:4865–4874. doi: 10.1128/JB.187.14.4865-4874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson MR, Conners SB, Montero CI, Chou CJ, Shockley KR, Kelly RM. The Thermotoga maritima phenotype is impacted by syntrophic interaction with Methanococcus jannaschii in hyperthermophilic coculture. Appl. Environ. Microbiol. 2006;72:811–818. doi: 10.1128/AEM.72.1.811-818.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maltsev N, Glass E, Sulakhe D, Rodriguez A, Syed MH, Bompada T, Zhang Y, D'S;ouza M. PUMA2—grid-based high-throughput analysis of genomes and metabolic pathways. Nucleic Acids Res. 2006;34:D369–D372. doi: 10.1093/nar/gkj095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poole FL, 2nd, Gerwe BA, Hopkins RC, Schut GJ, Weinberg MV, Jenney FE, Jr, Adams MW. Defining genes in the genome of the hyperthermophilic archaeon Pyrococcus furiosus: implications for all microbial genomes. J. Bacteriol. 2005;187:7325–7332. doi: 10.1128/JB.187.21.7325-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuijffel DF, van Empel PC, Pennings AM, van Putten JP, Nuijten PJ. Successful selection of cross-protective vaccine candidates for Ornithobacterium rhinotracheale infection. Infect. Immun. 2005;73:6812–6821. doi: 10.1128/IAI.73.10.6812-6821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang Z, Zheng W, He Y. BBP: Brucella genome annotation with literature mining and curation. BMC Bioinformatics. 2006;7:347. doi: 10.1186/1471-2105-7-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahren D, Tsoka S, Darzentas N, Kunin V, Lopez-Bigas N. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]