Abstract
Peroxisomes are essential organelles that play a key role in redox signalling and lipid homeostasis. They contain a highly diverse enzymatic network among different species, mirroring the varied metabolic needs of the organisms. The previous PeroxisomeDB version organized the peroxisomal proteome of humans and Saccharomyces cerevisiae based on genetic and functional information into metabolic categories with a special focus on peroxisomal disease. The new release (http://www.peroxisomeDB.org) adds peroxisomal proteins from 35 newly sequenced eukaryotic genomes including fungi, yeasts, plants and lower eukaryotes. We searched these genomes for a core ensemble of 139 peroxisomal protein families and identified 2706 putative peroxisomal protein homologs. Approximately 37% of the identified homologs contained putative peroxisome targeting signals (PTS). To help develop understanding of the evolutionary relationships among peroxisomal proteins, the new database includes phylogenetic trees for 2386 of the peroxisomal proteins. Additional new features are provided, such as a tool to capture kinetic information from Brenda, CheBI and Sabio-RK databases and more than 1400 selected bibliographic references. PeroxisomeDB 2.0 is a freely available, highly interactive functional genomics platform that offers an extensive view on the peroxisomal metabolome across lineages, thus facilitating comparative genomics and systems analysis of the organelle.
INTRODUCTION
Peroxisomes were discovered in 1954 with electron microscopy (1) and isolated by De Duve and Baudhuin in 1966 (2). Only recently, their evolutionary and ontogenetic origin has been related to the endoplasmic reticulum (3–5). Peroxisomes contribute to many crucial metabolic processes such as fatty acid oxidation, biosynthesis of ether lipids and free radical detoxification (6–8). These organelles are essential for normal development and growth, and their deficiencies lead to severe and often fatal inherited peroxisomal disorders in humans (9). With the aim of gaining knowledge of peroxisome biology, we have created PeroxisomeDB (http://www.peroxisomeDB.org), a relational database that compiles and integrates functional and genomic information about peroxisomal proteomes of Homo sapiens and Saccharomyces cerevisiae, organized in metabolic pathways (10). Key features of the database included a peroxisomal targeting signal (PTS) predictor and a peroxisome predictor, useful to identify peroxisomes and candidate peroxisomal proteins in newly sequenced genomes (10).
Peroxisomes differ substantially among species with respect to their enzyme content and specific metabolic functions. Some examples of the remarkable metabolic plasticity of peroxisomes include: (i) photorespiration in photosynthetic organisms; (ii) conversion of fatty acids into carbohydrates through the glyoxylate cycle in plants, protozoa and some yeasts (therefore designated as glyoxysomes); and (iii) glycolysis, purine salvage and pyrimidine biosynthesis, and pentose phosphate pathway in the glycosomes of trypanosomatids (11–13). To better compile the diversity of peroxisomes, we have introduced PeroxisomeDB 2.0. We generated this comprehensive catalogue by searching 37 taxonomically diverse full-sequenced genomes and identified the global peroxisomal metabolome. The database includes high-quality trees to make it easier to understand the evolutionary history of peroxisome proteins. PeroxisomeDB 2.0 aims at organising curated and updated, biochemical, genomic and phylogenetic information on peroxisomal proteomes and their interactions.
RESULTS AND DISCUSSION
Capturing the peroxisomal proteome from 37 eukaryotic genomes
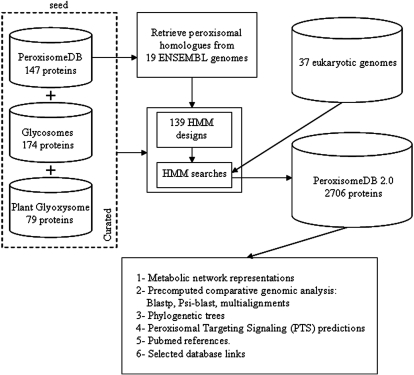
With a comparative genomics approach, we predicted peroxisomal proteins in silico based on homology (Figure 1). First, we obtained 85 H. sapiens and 61 S. cerevisiae peroxisomal proteins from PeroxisomeDB (10), the proteomes of glycosomes from the trypanosomatids Trypanosoma brucei, Trypanosoma cruzi and Leishmania major, manually curated according to literature (14), and the glyoxysomal proteome from the Arabidopsis thaliana genome contained in the Araperox database (15). Second, to identify peroxisomal protein homologs from 19 additional eukaryotic genomes in ENSEMBL, we used orthology predictions from ENSEMBL COMPARA (16) database version 47 (http://oct2007.archive.ensembl.org/). Third, from the resulting peroxisomal ensemble, we designed 139 peroxisomal protein families or core functional units, using a tool based on the profile Hidden Markov Models, HMMER (17). Finally, to identify the homologs to these peroxisomal protein family units, we calculated a HMM best hit score for searches against 37 eukaryotic genomes (35 new genomes, H. sapiens and S. cerevisiae). A putative peroxisomal homolog had an HMM score of at least 10−3. This search yielded a set of 2706 predicted peroxisomal homologous proteins, which were deposited into PeroxisomeDB 2.0.
Figure 1.
An outline on how the PeroxisomeDB 2.0 was generated. The PeroxisomeDB 2.0 includes proteins from yeast and human (PeroxisomeDB previous release), the glycosome proteome from trypanosomatids and the glyoxysome proteome from Arabidopsis thaliana. Using a protein consensus designed by HMM, we have scanned 37 eukaryotic genomes to identify 2706 putative peroxisomal homologs.
We sampled genomes with the goal of encompassing eukaryotic phylogenetic diversity, from mammals to protists or lower eukaryotes, including plants and fungi. The 37 eukaryotes included trypanosomatids (L. major, T. brucei and T. cruzi); the red algae Cyanidioschyzon merolae, a unicellular organism that contains a single peroxisome; the diatom Thalassiosira pseudonana, which acquired a plastid through a secondary endosymbiotic process; the ciliate Tetrahymena thermophila which is closely related to the Apicomplexa that have no peroxisomes (4,18); and the social amoeba Dictyostelium discoideum from a eukaryotic lineage prior to the split of metazoans and yeasts. A list of all species included in the database, and their genome sources is available at PeroxisomeDB 2.0 (http://www.peroxisomedb.org/show.php?action=OrganismCatalogue).
Fishing for PTS in silico
Our previously developed PTS predictor (10) is a powerful tool that provides PTS1, PTS2 and PEX19BS predictions by using a sequence multiple alignment without gaps or BLOCK (19). We performed high-scale motif searches using Blimps software (20) applied to each PTS BLOCK to screen the 2706 identified peroxisomal proteins. We identified 1126 PTSs from 944 proteins of the 2706 entries. Most proteins had either a PTS1 signal (571) or a PEX19BS signal (482). Only 73 proteins had PTS2 signals, although this motif is only known to be a part of 3-ketoacyl-CoA thiolase (Acaa1), Phytanoyl-CoA dioxygenase (Phyh) and Alkyldihydroxyacetonephosphate synthase (Agps) protein families. We confirmed a PTS2 signal in several known peroxisomal enzymes, such as Acyl-coenzyme A oxidase (Acox) and Citrate synthase (Cs) of D. discoideum and A. thaliana (21,22), and the Glycerol-3-phosphate dehydrogenase (GPD) of yeasts (23). We also predicted a PTS2 in the Glycosyl hydrolase (PEN2) of A. thaliana, experimentally identified as peroxisomal protein (24).
The global peroxisomal metabolome at a glance
Each of the 2706 proteins has a database entry that includes information on the assigned protein family, functional category and subcellular localisation, as well as links to reference databases and Pubmed. The entry also summarizes the PTS predictions and homology computations, including BLASTP, PSI-BLAST, multialignments and phylogenetic trees. Finally, enzymes entries are linked through the ontology-based mediator used by SBMM Assistant (www.sbmm.uma.es) (25) to the reference databases Brenda, CheBI and Sabio-RK, which provide updated kinetic and biochemical information for peroxisomal enzymes. This assistant works by recognizing the respective EC codes from Gene Bank and/or Ensemble and subsequently fetches the relevant data. A summary of the novel functions of PeroxisomeDB 2.0 as compared to the previous PeroxisomeDB is available as Supplementary Table S1.
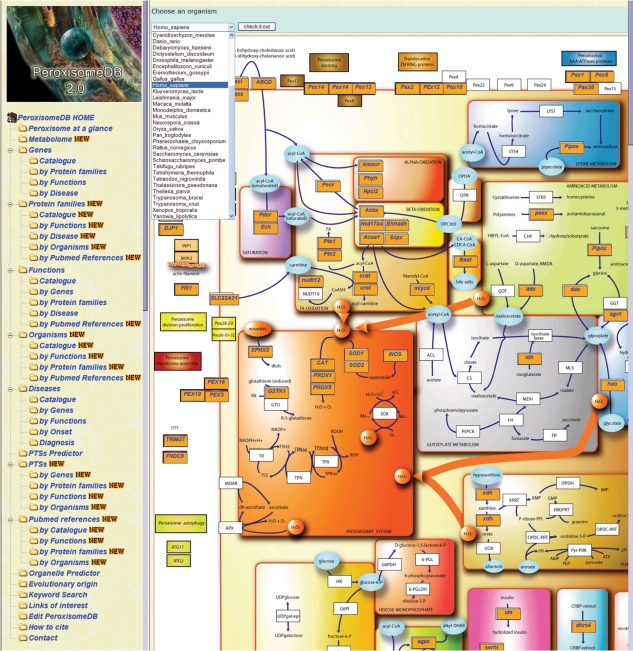
A central feature of the database is a global Peroxisome Metabolome Network composed of the most relevant known peroxisomal metabolic pathways, and it includes the putative peroxisomal homologs assigned to their respective protein families after identification in the current analysis (Figure 2). The integration into this comprehensive and complete metabolomic picture is of particular relevance for organisms such as Anopheles gambiae and Tetraodon nigroviridis, to cite but two, whose proteins lack curated peroxisomal annotation at NCBI and, to the best of our knowledge, experimental peroxisomal localisation (see Supplementary Table S2). We have developed a function for viewing the metabolome networks in a species-specific manner (Figure 2). Beyond these species-specific metabolic pathways, PeroxisomeDB 2.0 emerges as the integrative tool for the study of the global peroxisomal metabolome.
Figure 2.
The Metabolome page displays the global peroxisomal metabolome network for the 37 eukaryotic genomes. The tool allows the user to focus on a species of interest by clicking on ‘choose an organism’.
Molecular phylogenetic analysis of the global peroxisomal metabolome
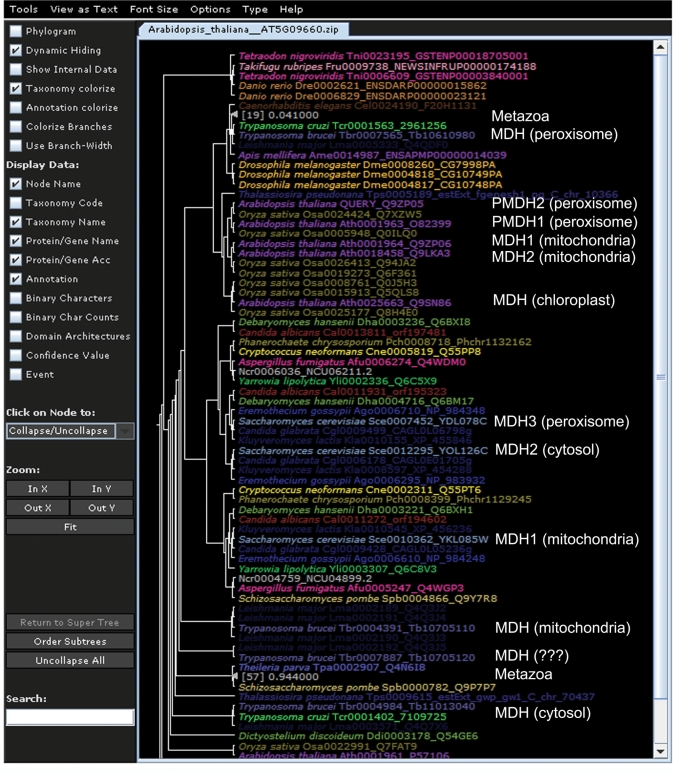
Using MUSCLE 3.6 (26), TrimAl v1.2 (27), PhyML aLRT version (28,29) and Mr Bayes (30) as described in PeroxisomeDB (http://www.peroxisomedb.org/peroxHelp/PROGRAMS_3.php), we built 2386 phylogenetic trees. An example of a phylogenetic tree for the Malate dehydrogenase (Mdh) enzyme is shown in Figure 3. For the remaining 320 peroxisomal homologs, we did not build a tree because fewer than four homologs were identified in the eukaryotic genomes. For some model species, such as S. cerevisiae and humans, links to additional phylogenomic resources are available through PhylomeDB database (31).
Figure 3.
The maximum likelihood tree (ML) of malate dehydrogenase (Mdh) protein family, taking the peroxisomal Mdh from Arabidopsis thaliana as a seed. Different species names are given in different colours. Phylogenetic trees are available in newick standard format and associated with taxonomic data via phyloXML, an XML language for the analysis, exchange, and storage of phylogenetic trees and associated data. The Archaeopteryx applet software was used for the visualisation of phylogenetic trees (37).
This molecular phylogenetic approach yielded important findings with functional implications. Beyond the well-known peroxisomal core metabolic pathways that confer identity to the organelle, such as catalase and beta-oxidation of fatty acids (6–8), we identified additional ancient peroxisomal acquisitions in taxonomically diverse organisms. For example, Peroxiredoxin 1 (Prdx1) in mammals (32) and Tryparedoxin peroxidase (Tpx) of trypanosomatids were closely related, suggesting a common peroxisomal origin for the peroxiredoxin system. Similarly, the trypanosomatid NADP+-dependent dehydrogenase Glucose-6-phosphate 1-dehydrogenase (G6pdh) of the pentose phosphate pathway of glycosomes is present in all eukaryotic organisms analysed here, although the peroxisomal localisation of the pathway has only been described in trypanosomatids to date. This finding might provide a rationale for previous experimental data showing 10% enzymatic activity of these enzymes in rat liver peroxisomes (33), raising the possibility of a partial peroxisomal location of the pathway in mammals. Additionally, the phylogenies revealed a common origin for metazoan Coenzyme A diphosphatase or nucleoside diphosphate-linked moiety X motif 7 (Nudt7 or Nudix motif 7) and the Peroxisomal nudix pyrophosphatase with specificity for coenzyme A and CoA derivatives (Pcd1) of yeasts, reinforcing previous suggestions (34).
Not all currently known peroxisomal enzymes were part of the original peroxisome, examples being some enzymes of beta-oxidation of fatty acids route (3). Peroxisomes of different lineages have also acquired enzymes as the metabolic needs of the organisms changed. Interestingly, we detected independent events of the same homologous protein from different species recruited into the peroxisome. Some enzymes of the glyoxylate pathway have been independently incorporated into the peroxisome of yeasts, plants and trypanosomatids (35). This is the case for the Mdh (see tree Figure 3), which evolved from duplications of homologous proteins in another cellular location, which occurred after eukaryote divergence. For Cs, duplications occurred in yeasts, but a horizontal transfer from eubacteria was at the origin of the peroxisomal enzyme in plants (35). This is an excellent example of the metabolic constraints of the cell acting as the driving force in the recruitment of a given key function to peroxisomes, several times independently.
Future challenges
As organelle-specific proteomic technologies and whole genome sequencing projects progress, the number of known peroxisomal proteins is exponentially growing. It is challenging to keep up with these protein discoveries while maintaining a high-quality curated database. The PeroxisomeDB 2.0 currently includes manually curated human, S. cerevisiae, A. thaliana and trypanosomatid peroxisomal proteins with the peroxisomal proteins from the remaining species assigned based on an automatic sequence homology. We plan to replace this predicted peroxisomal annotation to fetch experimental organelle localisation with a semi-automatic curation system for semantic data integration and text mining from literature, which will be relaunched every two months. Precomputed homology searches using BLAST and PSI-BLAST will also be relaunched bimonthly. A collaborative interface will be developed to encourage user input to complement these routines and benefit from the collective knowledge of the scientific community. Finally, peroxisomes are dynamic compartments metabolically linked to other subcellular structures, such as mitochondria and endoplasmic reticulum (36). Extension of the peroxisomal metabolome beyond the organelle borders to encompass these shared metabolic pathways is an exciting perspective.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant FIS PI080991 and 2009SGR85 (to A.P.); SAF2008-2522 (to F.S.-J.); Grant CVI-2999 (to A.R.-C.); FIS (ECA07/055 to A.S.). The work was developed under the COST action BM0604 (to A.P.). CIBERER is an initiative of the Instituto de Salud Carlos III. Funding for open access charge: Grant (FIS PI080991).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the computer resources, technical expertise and assistance provided by the Barcelona Supercomputing Center—Centro Nacional de Supercomputación, and The Spanish National Bioinformatics Institute—Instituto Nacional de Bioinformática (INB). Part of the simulations have been done at the freely available Bioportal (www.bioportal.uio.no).
REFERENCES
- 1. Rhodin (1954) Correlation of ultrastructural organization and function in normal experimentally changed convoluted tubule cells of the mouse kidney. Thesis. Karolinska Institutet, Stockholm, Aktiebolaget Godvil.
- 2.De Duve C, Baudhuin P. Peroxisomes (microbodies and related particles) Physiol. Rev. 1966;46:323–357. doi: 10.1152/physrev.1966.46.2.323. [DOI] [PubMed] [Google Scholar]
- 3.Gabaldon T, Snel B, van Zimmeren F, Hemrika W, Tabak H, Huynen MA. Origin and evolution of the peroxisomal proteome. Biol. Direct. 2006;1:8. doi: 10.1186/1745-6150-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schluter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A. The evolutionary origin of peroxisomes: an ER-Peroxisome Connection. Mol. Biol. Evol. 2006;23:838–845. doi: 10.1093/molbev/msj103. [DOI] [PubMed] [Google Scholar]
- 5.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 6.Bonekamp NA, Volkl A, Fahimi HD, Schrader M. Reactive oxygen species and peroxisomes: struggling for balance. Biofactors. 2009;35:346–355. doi: 10.1002/biof.48. [DOI] [PubMed] [Google Scholar]
- 7.Schrader M, Fahimi HD. The peroxisome: still a mysterious organelle. Histochem. Cell Biol. 2008;129:421–440. doi: 10.1007/s00418-008-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 9.Wanders RJ. Metabolic and molecular basis of peroxisomal disorders: a review. Am. J. Med. Genet. 2004;126A:355–375. doi: 10.1002/ajmg.a.20661. [DOI] [PubMed] [Google Scholar]
- 10.Schluter A, Fourcade S, Domenech-Estevez E, Gabaldon T, Huerta-Cepas J, Berthommier G, Ripp R, Wanders RJ, Poch O, Pujol A. PeroxisomeDB: a database for the peroxisomal proteome, functional genomics and disease. Nucleic Acids Res. 2007;35:D815–D822. doi: 10.1093/nar/gkl935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunze M, Pracharoenwattana I, Smith SM, Hartig A. A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim. Biophys. Acta. 2006;1763:1441–1452. doi: 10.1016/j.bbamcr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Michels PA, Bringaud F, Herman M, Hannaert V. Metabolic functions of glycosomes in trypanosomatids. Biochim. Biophys. Acta. 2006;1763:1463–1477. doi: 10.1016/j.bbamcr.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Reumann S, Weber AP. Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled—others remain. Biochim. Biophys. Acta. 2006;1763:1496–1510. doi: 10.1016/j.bbamcr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Opperdoes FR, Szikora JP. In silico prediction of the glycosomal enzymes of Leishmania major and trypanosomes. Mol. Biochem. Parasitol. 2006;147:193–206. doi: 10.1016/j.molbiopara.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Reumann S, Ma C, Lemke S, Babujee L. AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 2004;136:2587–2608. doi: 10.1104/pp.104.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogh A, Brown M, Mian IS, Sjolander K, Haussler D. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 18.Ding M, Clayton C, Soldati D. Toxoplasma gondii catalase: are there peroxisomes in toxoplasma? J. Cell Sci. 2000;113(Pt 13):2409–2419. doi: 10.1242/jcs.113.13.2409. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Blocks-based methods for detecting protein homology. Electrophoresis. 2000;21:1700–1706. doi: 10.1002/(SICI)1522-2683(20000501)21:9<1700::AID-ELPS1700>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 21.Huang YC, Chen YH, Lo SR, Liu CI, Wang CW, Chang WT. Disruption of the peroxisomal citrate synthase CshA affects cell growth and multicellular development in Dictyostelium discoideum. Mol. Microbiol. 2004;53:81–91. doi: 10.1111/j.1365-2958.2004.04109.x. [DOI] [PubMed] [Google Scholar]
- 22.Pracharoenwattana I, Cornah JE, Smith SM. Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell. 2005;17:2037–2048. doi: 10.1105/tpc.105.031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valadi A, Granath K, Gustafsson L, Adler L. Distinct intracellular localization of Gpd1p and Gpd2p, the two yeast isoforms of NAD+-dependent glycerol-3-phosphate dehydrogenase, explains their different contributions to redox-driven glycerol production. J. Biol. Chem. 2004;279:39677–39685. doi: 10.1074/jbc.M403310200. [DOI] [PubMed] [Google Scholar]
- 24.Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 25.Reyes-Palomares A, Montanez R, Real-Chicharro A, Chniber O, Kerzazi A, Navas-Delgado I, Medina MA, Aldana-Montes JF, Sanchez-Jimenez F. Systems biology metabolic modeling assistant: an ontology-based tool for the integration of metabolic data in kinetic modeling. Bioinformatics. 2009;25:834–835. doi: 10.1093/bioinformatics/btp061. [DOI] [PubMed] [Google Scholar]
- 26.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 29.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 30.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 31.Huerta-Cepas J, Bueno A, Dopazo J, Gabaldon T. PhylomeDB: a database for genome-wide collections of gene phylogenies. Nucleic Acids Res. 2008;36:D491–D496. doi: 10.1093/nar/gkm899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Immenschuh S, Baumgart-Vogt E, Tan M, Iwahara S, Ramadori G, Fahimi HD. Differential cellular and subcellular localization of heme-binding protein 23/peroxiredoxin I and heme oxygenase-1 in rat liver. J. Histochem. Cytochem. 2003;51:1621–1631. doi: 10.1177/002215540305101206. [DOI] [PubMed] [Google Scholar]
- 33.Antonenkov VD. Dehydrogenases of the pentose phosphate pathway in rat liver peroxisomes. Eur. J. Biochem. 1989;183:75–82. doi: 10.1111/j.1432-1033.1989.tb14898.x. [DOI] [PubMed] [Google Scholar]
- 34.Gasmi L, McLennan AG. The mouse Nudt7 gene encodes a peroxisomal nudix hydrolase specific for coenzyme A and its derivatives. Biochem. J. 2001;357:33–38. doi: 10.1042/0264-6021:3570033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnarrenberger C, Martin W. Evolution of the enzymes of the citric acid cycle and the glyoxylate cycle of higher plants. A case study of endosymbiotic gene transfer. Eur. J. Biochem. 2002;269:868–883. doi: 10.1046/j.0014-2956.2001.02722.x. [DOI] [PubMed] [Google Scholar]
- 36.Schrader M, Yoon Y. Mitochondria and peroxisomes: are the ‘big brother' and the ‘little sister' closer than assumed? Bioessays. 2007;29:1105–1114. doi: 10.1002/bies.20659. [DOI] [PubMed] [Google Scholar]
- 37.Zmasek CM, Eddy SR. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]