Abstract
The Saccharomyces Genome Database (SGD; http://www.yeastgenome.org) is a scientific database for the molecular biology and genetics of the yeast Saccharomyces cerevisiae, which is commonly known as baker’s or budding yeast. The information in SGD includes functional annotations, mapping and sequence information, protein domains and structure, expression data, mutant phenotypes, physical and genetic interactions and the primary literature from which these data are derived. Here we describe how published phenotypes and genetic interaction data are annotated and displayed in SGD.
INTRODUCTION
The Saccharomyces Genome Database (SGD; http://www.yeastgenome.org) collects, organizes and makes scientific information regarding the genes, proteins and other chromosomal features of the model organism Saccharomyces cerevisiae freely available to the public. This information includes descriptions of phenotypes that arise from mutations in single genes, or from genetic interactions between two mutated genes. At SGD, results from traditional bench experiments published in the scientific literature have been the primary source of phenotype annotations, and a number of large-scale studies have greatly increased the available phenotype data. The integration of these phenotypic and genetic interaction data into SGD in a comprehensive and coherent manner has been a major focus of our recent activities because they can help users uncover new function or process information, particularly for genes that are not well characterized.
Phenotypes are observable characteristics of an organism that arise due to the interaction of its genotype with its environment. A mutant phenotype is an altered attribute resulting from changes in the genome. A mutant yeast phenotype can be any detectable feature of cells, colonies or cultures. Curated phenotypes in SGD include morphological, developmental or growth-related manifestations of mutations in single genes, observable at the cellular level in living cells. Some molecular phenotypes, such as aberrant protein accumulation or chemical compound excretion, are also captured if they occur in vivo. Mutant phenotypes inferred from assays performed in vitro or in organello are not recorded (1). Curated genetic interactions in SGD describe the ways in which concurrent mutations in two separate genes can interact with each other, including suppression, complementation and synthetic lethality. Data from large-scale screens for genetic interactions, such as those from Synthetic Genetic Analysis, Diploid-based Synthetic Lethality Analysis on Microarrays or other types of genetic interaction screens that use genomic techniques, are also included.
SINGLE MUTANT PHENOTYPES
SGD curators have conducted focused surveys of the yeast literature to find and record mutant phenotypes for every gene in the yeast genome. This sweep included many pivotal early papers that characterized classical yeast phenotypes and identified the genes involved, and has also included recent large-scale studies, some of which report mutant phenotypes for hundreds or even thousands of genes at a time. In addition to these targeted phenotype curation efforts, SGD curators record phenotypes from newly published papers on a weekly basis as part of regular curation activity. Note that in order to keep the data tractable, only ‘representative’ phenotypes are annotated; SGD does not curate every possible phenotype from every paper.
Mutant phenotype data are recorded in the database using controlled vocabularies. The mutant phenotypes themselves are each recorded with an ‘observable’ and a ‘qualifier’. The observable is the main feature of the phenotype, whereas the qualifier describes the direction or type of change relative to wild type. Qualifiers include the following: absent, arrested, delayed, premature, abnormal, decreased, decreased duration, decreased rate, increased, increased duration, increased rate, normal, normal duration and normal rate. Observables are organized as an ontology of terms describing cellular processes such as the cell cycle, intracellular transport and stress or chemical resistance; developmental processes such as budding, mating and sporulation; metabolic processes such as growth and nutrient utilization; and other characteristics such as cellular morphology or culture appearance. A complete hierarchical list of current observables is available at http://www.yeastgenome.org/cache/PhenotypeTree.html. Clicking on any term on this page leads to a list of all phenotypes annotated using this term, as well as the genes associated with them. The phenotype terms in use at SGD are also being used by other fungal databases, such as the Aspergillus Genome Database [AspGD; (2)]. The entire list of observables and qualifiers, including definitions and relationships between terms, is contained within the ‘Ascomycete phenotype ontology’, which can be downloaded from the Open Biomedical Ontologies Foundry (http://www.obofoundry.org/), an open-access collection of scientific controlled vocabularies (3).
Controlled vocabularies are also used to describe additional aspects of the phenotype, including details about the causal mutation (such as mutant type), the conditions of its occurrence (such as strain background) and the assay performed to elucidate its effects (experiment type, chemicals, etc.). Mutant types include the following classes: activation, conditional, dominant negative, gain of function, reduction of function, null (i.e. ‘loss of function’), misexpression, overexpression, repressible (i.e. ‘depletion’) and unspecified. ‘Activation’ refers to a mutation that increases the normal activity of the gene product. ‘Conditional’ indicates that the phenotype appears only under certain conditions (e.g. elevated temperature). ‘Dominant negative’ signifies that the mutant gene product negatively affects the activity of the wild-type gene product, such as by dimerizing with it or titrating one of its targets. ‘Gain of function’ denotes a mutation that confers new activity on the gene product. ‘Reduction of function’ represents a mutation that reduces the activity of the gene product. ‘Null’ designates loss of a gene product’s activity, and can be used to describe complete deletions of a gene, as well as point mutations in key residues.
Allele names and strain backgrounds are noted when that information is available in the publication from which the phenotype is being curated. Descriptions are included for allele names when possible, such as ‘tim9-1 (G71R)’. If alleles are not given formal names in the publication, but instead reported only as amino acid substitutions, then the amino acid substitution is incorporated as the allele name (e.g. ‘tim9-E52K’). Allele names that have been reported using alternative gene names are combined with the standard SGD gene name [e.g. ‘tom22-(mas22-4)’]. Strain backgrounds are recorded to document the genetic environment in which the mutant phenotype was characterized. Only the most commonly used strain backgrounds are entered: CEN.PK, D273-10B, FL100, JK9-3d, RM11-1a, S288C, SEY6210, SK1, Sigma1278b, W303, X2180-1A and Y55. Strain backgrounds not included in this list are displayed as ‘Other’. The SGD Wiki includes information about these strain backgrounds, including published references (see http://wiki.yeastgenome.org/index.php/Commonly_used_strains).
The assays performed to detect and analyze the mutant phenotypes are termed ‘experiment types’ and fall into two major categories: classical genetics and large-scale survey. Small-scale experiments that focus on one or a few genes are recorded as ‘classical genetics’. ‘Large-scale survey’ denotes experiments that have been designed typically with some knowledge of the genome sequence, such as systematic mutation sets that include collections of deletion strains. Large-scale surveys also comprise those that use high-throughput, robot-assisted techniques, such as competitive growth assays in which pools of mutant strains are grown together for many generations to assess relative fitness. For all experiment types, haploidy is implied unless otherwise noted. The use of homozygous or heterozygous diploid strains in an experiment is explicitly stated when relevant.
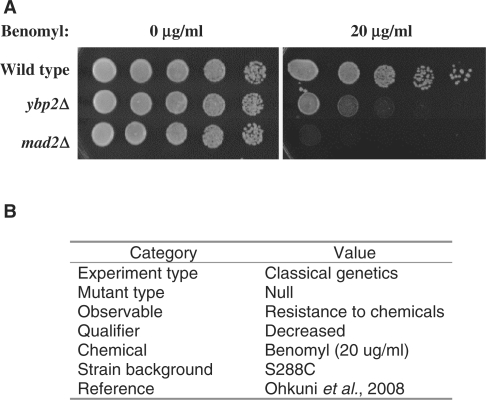
Chemical compounds used in a phenotype assay, such as nutrient sources or exogenous chemicals that affect growth, are recorded using controlled vocabulary terms from the Chemical Entities of Biological Interest database maintained at the European Bioinformatics Institute (ChEBI; http://www.ebi.ac.uk/chebi/; 4). SGD curators contribute to the development of ChEBI by suggesting new terms when necessary. Other pertinent details are also documented, such as experimental conditions under which the phenotype is observed (e.g. growth media or temperature), any reporters used or any other facts that might be helpful for understanding the phenotype being reported. All curated phenotypes also cite the publication in which the phenotype is described. Some of these properties of a phenotype annotation are illustrated in Figure 1. The assay shown is curated for both YBP2 and MAD2 using experiment type ‘classical genetics’, mutant type ‘null’, observable ‘resistance to chemicals’, qualifier ‘decreased’ and chemical ‘benomyl’ with detail ‘20 μg/ml’. Also included in the phenotype annotation are the strain background, which in this case is S288C, and the reference from which this phenotype was curated (5).
Figure 1.
The ybp2 and mad2 phenotypes represented by the growth assay shown in (A) are each curated in SGD as listed in (B).
Phenotype data are displayed on locus-specific pages that list all curated phenotypes for a particular gene. These pages are accessible via the ‘Phenotype’ tab on the Locus Summary and also from the Mutant Phenotypes section of the Locus Summary, where the phenotype data are presented in summary form. Data are presented in tabular format on the Phenotypes page, and may continue onto additional pages if the number of curated phenotypes exceeds 30. All of the phenotype data for a particular gene may be downloaded as a tab-delimited text file through the ‘Download Data’ links near the top and bottom of the Phenotypes page. A file containing phenotype data for all genes is available for download at http://downloads.yeastgenome.org/pub/yeast/literature_curation/phenotype_data.tab.
Phenotype data may be searched either via the SGD Search, which can be accessed from the search box located at the top of most SGD pages and queries the ‘observable’ terms, or through the Expanded Phenotype Search on the Search Options page, which offers a complete search of phenotypes and associated data. For the SGD Search, text entered is used to search the list of phenotype terms describing observed features (observables), as well as many other kinds of data in SGD. The results page lists the matches found in various types of data. A typical search result for phenotypes would appear as ‘12 Phenotype annotations [Expanded Phenotype Search]’, where the phrase ‘12 Phenotype annotations’ is hyperlinked to a list of phenotypes for which the search criterion matches all or part of a phenotype term. The ‘Expanded Phenotype Search’ link is hyperlinked to a list of search results in which the original search criterion has been used to search all phenotype data, including the terms describing observable features and other associated information such as details, conditions and strain names. The ‘Analyze Gene List’ link at the top of every phenotype search results page leads to a section at the bottom of the page that provides links to tools for further analysis of the listed genes and options for downloading the displayed information: GO Term Finder, GO Slim Mapper, View GO Annotation Summary, Download Data and Download options (6). The ‘Advanced Search’ interface, which is accessible via a link in the toolbar running across the top of most SGD web pages, can also be used to query chromosomal features using multiple criteria, including a limited phenotype search for genes annotated as ‘viable’ or ‘inviable’ in systematic deletion experiments. These searches will display a list of genes that when mutated share a common phenotype, and can be further analyzed to determine if the genes also share common cellular roles.
PHENOTYPES FROM GENETIC INTERACTIONS
Phenotypes resulting from mutations in more than one gene are captured as genetic interactions. All of the curated interaction data in SGD are loaded in bulk from the BioGRID database of physical and genetic interactions (Biological General Repository for Interaction Datasets; http://www.thebiogrid.org/) hosted by Mike Tyers’ group at the University of Toronto (7). BioGRID contains high-throughput interaction data for S. cerevisiae and other model organisms, and for yeast also contains interactions curated from small-scale studies. The curation of the BioGRID interaction data is a collaborative effort between the Tyers lab, the SGD Colony at Princeton University, and the SGD Project at Stanford University. After curation at BioGRID, the interaction data are transferred to SGD on a monthly basis.
Interaction data are recorded in BioGRID using a controlled vocabulary to describe the various types of experiments performed to assess genetic interactions, such as dosage lethality, growth defect or rescue; and synthetic lethality, growth defect or rescue. In SGD, genetic interactions that affect viability or growth are also described using the same controlled phenotype vocabulary used for single mutant phenotypes (Table 1). For example, genetic interactions curated at BioGRID as a ‘dosage growth defect’ are annotated in SGD using the observable ‘vegetative growth’ with qualifier ‘decreased’. The representation of phenotypes that arise from genetic interactions using the same vocabulary that describes single mutant phenotypes facilitates comparison between the two different types of phenotype data in SGD.
Table 1.
Genetic interactions captured at BioGRID that relate to growth or viability are annotated in SGD using a subset of the same controlled vocabulary terms used to describe single mutant phenotypes
| BioGRID experimental system | Phenotype annotation in SGD |
|---|---|
| Dosage lethality | Inviable |
| Dosage growth defect | Vegetative growth: decreased |
| Dosage rescue | Vegetative growth: normal |
| Synthetic lethality | Inviable |
| Synthetic growth defect | Vegetative growth: decreased |
| Synthetic rescue | Vegetative growth: normal |
Interaction data are displayed in tabular format on locus-specific Interactions pages, which are accessible via the ‘Interactions’ tab on the Locus Summary and also from the Interactions section of the Locus Summary, where the interaction data are presented in summary form. Data are presented on the Interactions pages in a configurable table that allows sorting and filtering of the data. The ‘Analyze Gene List’ module, as described earlier, is present at the bottom of the page, providing download options and facilitating further analysis of the listed genes. All of the interaction data for a particular gene can be downloaded as a tab-delimited text file via the ‘Download Unfiltered Data’ link at the bottom of the page. The ‘Advanced Search’ interface, as mentioned above, includes a limited interaction option for the inclusion of genes annotated with or without interaction data. A file containing interaction data for all genes is available for download at http://downloads.yeastgenome.org/pub/yeast/literature_curation/interaction_data.tab.
SUMMARY
The SGD Project’s goal in presenting phenotype data reflects its central mission to aid researchers in sorting through large amounts of data, juxtaposing relevant pieces of information that they might otherwise have missed. SGD is the central resource for yeast mutant phenotypes. The published yeast literature is curated to provide representative phenotype annotation, and controlled vocabularies are used to describe the observable characteristics for each mutant phenotype. SGD is dedicated to maintain curated data of the utmost quality and is receptive to questions and comments. Please contact us at yeast-curator@yeastgenome.org.
FUNDING
National Human Genome Research Institute (HG001315 to J.M.C.). Funding for open access charge: National Human Genome Research Institute.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The image used in Figure 1 is from Ohkuni et al. (5), and is being used under the Creative Commons Attribution License (CCAL; http://creativecommons.org/licenses/by/2.5/).
REFERENCES
- 1.Costanzo MC, Skrzypek MS, Nash R, Wong E, Binkley G, Engel SR, Hitz B, Hong EL, Cherry JM the Saccharomyces Genome Database Project. New mutant phenotype data curation system in the Saccharomyces Genome Database. Database. 2009 doi: 10.1093/database/bap001. doi: 10.1093/database/bap001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaud MB, Chibucos MC, Costanzo MC, Crabtree J, Inglis DO, Lotia A, Orvis J, Shah P, Skrzypek MS, Binkley G, et al. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res. 2009;37:D420–D427. doi: 10.1093/nar/gkp751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotech. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohkuni K, Abdulle R, Tong AHY, Boone C, Kitagawa K. Ybp2 associates with the central kinetochore of Saccharomyces cerevisiae and mediates proper mitotic progression. PLoS ONE. 2008;3:e1617. doi: 10.1371/journal.pone.0001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bähler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]