Abstract
The European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) is Europe’s primary nucleotide sequence archival resource, safeguarding open nucleotide data access, engaging in worldwide collaborative data exchange and integrating with the scientific publication process. ENA has made significant contributions to the collaborative nucleotide archival arena as an active proponent of extending the traditional collaboration to cover capillary and next-generation sequencing information. We have continued to co-develop data and metadata representation formats with our collaborators for both data exchange and public data dissemination. In addition to the DDBJ/EMBL/GenBank feature table format, we share metadata formats for capillary and next-generation sequencing traces and are using and contributing to the NCBI SRA Toolkit for the long-term storage of the next-generation sequence traces. During the course of 2009, ENA has significantly improved sequence submission, search and access functionalities provided at EMBL–EBI. In this article, we briefly describe the content and scope of our archive and introduce major improvements to our services.
BRIEF HISTORY
ENA was established in the early 1980s as the EMBL Data Library (later renamed as the EMBL Nucleotide Sequence Database, EMBL-Bank) and focused initially on richly annotated nucleotide sequences. After breakthrough improvements in sequencing technologies culminating in the wide-scale adoption of the chain-termination method developed by Sanger (1,2), a further function of the archive, initially operated by the Wellcome Trust Sanger Institute as the Trace Archive, was the storage of high-throughput sequence reads with associated quality and instrumentation information. The growth of the Trace Archive accelerated notably with the emergence of the shotgun approach as the method of choice for genome sequencing and increased further with the commercialization of highly parallel next-generation sequencing technologies first by Roche’s 454 (http://www.454.com/) followed by Illumina’s Genome Analyzer (http://www.illumina.com/pages.ilmn?ID=204) and Applied Biosystems' SOLID System (http://www3.appliedbiosystems.com/AB_Home/applicationstechnologies/SOLiD-System-Sequencing-B/index.htm) (3). After inclusion of the Trace Archive and the establishment of the Sequence Read Archive (SRA) in 2008, an archival resource for next-generation sequences, ENA had completed its transformation into a comprehensive nucleotide sequence archive.
FREE AND UNRESTRICTED ACCESS
ENA, along with NCBI (4) and DDBJ (5), is an active member of the International Nucleotide Sequence Database Collaboration (INSDC), established to promote worldwide collaborative data exchange. The principal policy of INSDC is to provide free and unrestricted permanent access to all archived nucleotide data. All primary data in the INSDC belong to the submitters and can only be updated with submitter consent. For full policy details, please refer to http://www.insdc.org/page.php?page=policy.
STRUCTURE
The ENA consists of ENA-Annotation, ENA-Assembly, and ENA-Reads tiers. The oldest records lie within ENA-Annotation and ENA-Assembly sections (Table 1). Capillary and next-generation sequence traces are included in ENA-Reads (Table 2). Capillary traces are stored in the Trace Archive and next-generation sequences in the SRA. Different data classes are designed to capture the full spectrum of nucleotide-sequence-related information starting from the sequencing experiments through complete assemblies and annotations up to high-level sample and project information. ENA-Annotation contains rich high-level functional annotation captured in the INSDC feature table format. ENA-Assembly is designed for efficient storage of assembly information and ENA-Reads for the efficient storage of sequence trace information. Entries from different data classes are connected together through high-level sample and project records to create rich linkage between different types of data.
Table 1.
ENA-Annotation and ENA-Assembly data classes
| Data class | Description |
|---|---|
| Expressed sequence tag (EST) | High-throughput short transcribed cDNA (mRNA) sequences |
| Genome survey sequence (GSS) | High-throughput short genomic sequences |
| High throughput cDNA sequencing (HTC) | Unfinished cDNA (mRNA) sequences |
| High throughput genome sequencing (HTG) | Unfinished genomic sequences |
| Patent sequence (PAT) | Patent sequences |
| Sequenced tagged site (STS) | Short unique genomic sequences |
| Standard sequence (STD) | High-quality annotated sequences |
| Third party annotation sequence (TPA) | Re-annotated and re-assembled sequences |
| Transcriptome shotgun assembly (TSA) | Computationally assembled sequences |
| Whole genome shotgun (WGS) | Shotgun sequences |
| Constructed sequences (CON) | Sequence assemblies primarily from WGS sequences |
Table 2.
ENA-Reads data classes
| Data class | Description |
|---|---|
| Trace Archive | Sequence traces with base, quality and intensity information from capillary sequencing instruments |
| Sequence read archieve (SRA) | Sequence traces with base, quality and intensity information from next-generation sequencing instruments |
CONTENT
In October 2009, ENA-Annotation and ENA-Assembly contained 163 million records covering 283 billion bases. Whole-genome shotgun sequences continue to be the dominant source of new sequences (30% sequences and 53% of bases) followed by expressed sequence tags (EST) (38% sequences and 12% of bases). The growth of the Trace Archive, part of ENA-Reads, is markedly reduced, increasing only 6.2% in the last year to 1.96 billion sequences and 1.77 trillion bases. The SRA, containing next-generation sequences, has rapidly grown to 83 billion spots covering 7.4 trillion bases, making the SRA the fastest growing section of ENA. In ENA, the number of sequenced taxa has grown to 460 000 organisms and the number of scientific literature citations has exceeded 270 000.
IMPROVED INTERACTIVE SUBMISSION TOOL
We have made significant improvements to our interactive submission tool (Webin) with the addition of a new template-based system. Webin templates are text documents containing information common to large numbers of similar records and variable fields expected to be of use for a given data type. At the end of the submission process, submitted information is expanded using the template to create full database records. The Webin launcher, the entry point to all interactive submissions, has been extended to offer an appropriate set of common use case templates for submitters and to guide them through the submissions process.
Presently, we have configured templates for most commonly occurring types of ENA-Annotation submissions, including a MIENS (Minimum Information about an ENvironmental Sequence) standard compliant template, and we may add additional templates complying with other standards as they become available. We also plan to expand this system to cover SRA and project submissions. Upon submission and template expansion, the resulting entries are analysed with a rule-based validator and users are informed of any warnings and errors generated as part of the data validation process. All users wishing to submit large number of sequences with a fixed number of variable fields are encouraged to contact datasubs@ebi.ac.uk for creation of new templates which can be rapidly integrated into Webin. The Webin submission tool is available at http://www.ebi.ac.uk/embl/Submission/webin.html.
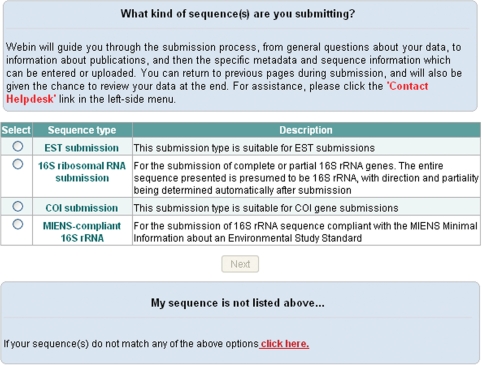
On the first page, users are asked to choose one of the available sequence submission types (Figure 1). This will determine which template will be used for submission.
Figure 1.
Selection of the sequence submission type.
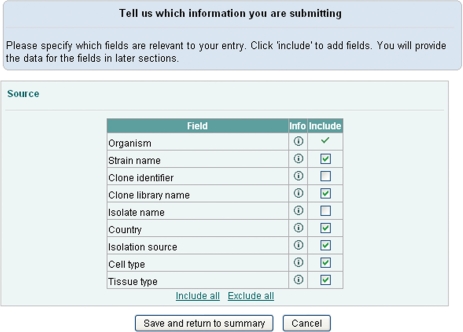
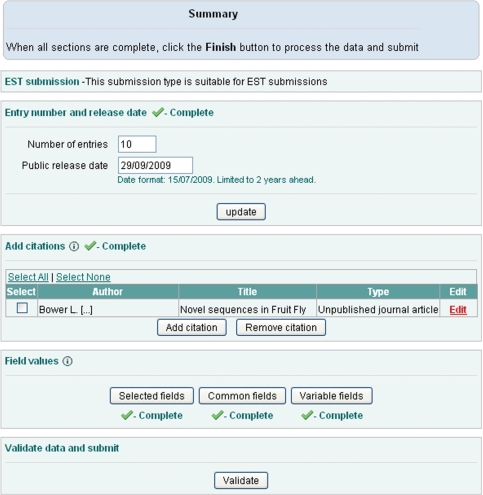
Our template-based submission tool supports both constant and variable parameters for templates. Parameters are selected on the second page from a list of mandatory and optional fields (Figure 2). Constant common parameters are selected and filled in on the third page and the variable parameters are uploaded on the fourth page using a comma separated text file. This file is generated by Webin for the user based on the variable field selection and contains one column for each variable field. It is expected to be filled up by the submitter, e.g. by using Excel, and to contain the information for each sequence on its own row. Finally, the summary page provides an overview of the progress of the submission (Figure 3). Data is validated using the ‘validate’ button after which it can be submitted to the archive. Curator assistance can be requested from most pages.
Figure 2.
Selection of the fields to include in the submission.
Figure 3.
Submission summary page.
SRA AUTOMATED SUBMISSION TOOL
The SRA accepts sequence submissions generated by the next-generation sequencing platforms. New submitters are advised to contact datasubs@ebi.ac.uk for the creation of a submission account and a secure data upload area. An automated submission service is provided to all registered submitters and is recommended for all users providing regular submissions. Immediate feedback is given of metadata validation errors and a service is provided for querying the data file processing status.
The first step in the submission process is to upload data files in platform specific, SRF or fastq formats using FTP or Aspera protocols into the secure data upload area. Aspera (http://www.asperasoft.com/) is a commercial UDP-based data transfer protocol capable of better utilization of available network bandwidth than the TCP-based FTP protocol.
The second step is the preparation of submission, study, sample, experiment and run SRA metadata XML files. Studies and samples contain high-level project and sample information. Each experiment is associated with a single study and one or more samples. Experiments contain one or more runs which are associated with the submitted data files. The final step is to use our RESTful web-based service (https://www.ebi.ac.uk/ena/submit/drop-box/) to submit the data files and the SRA XML objects. Interactive submissions use the submission form and fully automated submissions take advantage of the RESTful service.
ENA BROWSER
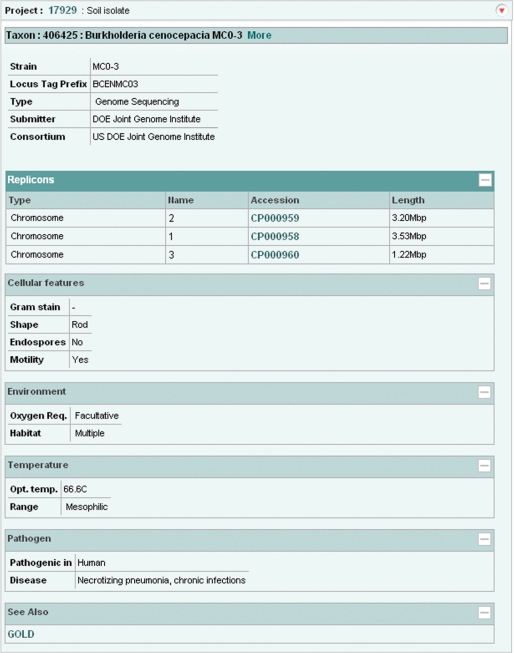
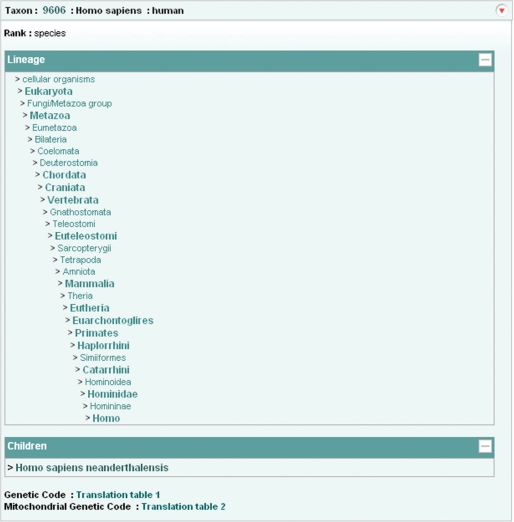
We have developed a new web-based data retrieval and visualization tool which has been first deployed for the SRA, Project and Taxonomy data, and which will soon be expanded to cover the remaining ENA-Reads data (from the Trace Archive) and ENA-Assembly and ENA-Annotation. Data can be visualized and downloaded in XML, HTML and flat file formats. Retrievals can be made by single accession numbers, e.g. http://www.ebi.ac.uk/ena/data/view/SRP000031&display=html, ranges of accession numbers, e.g. http://www.ebi.ac.uk/ena/data/view/ERX000025-ERX000034&display=html, or by lists of accession numbers, e.g. http://www.ebi.ac.uk/ena/data/view/ERR001087,ERR001088&display=html. Numeric project and taxonomy identifiers must be prefixed with ‘Project:’ and ‘Taxon:’, e.g. http://www.ebi.ac.uk/ena/data/view/Project:10724&display=html (Figure 4) and http://www.ebi.ac.uk/ena/data/view/Taxon:9606&display=html (Figure 5). Display in XML and HTML format is requested by using ‘display=xml’ and ‘display=html’ attributes, respectively. Download in gzip compressed format is possibly by using ‘download=gzip’ in place of ‘display’ attribute. SRA data can be downloaded either in submitted or fastq format by clicking links displayed in the SRA submission and run pages.
Figure 4.
ENA Browser project page.
Figure 5.
ENA Browser taxonomy page.
The ENA browser has been fully integrated with the EB-Eye indexer accessible from the header section of all EBI web pages. Users search on accession numbers, description text or other free text to find appropriate data in the ENA Browser.
ENA SEQUENCE SIMILARITY SEARCH
Early in 2010, we expect to launch a new sequence similarity search service based on Exonerate (6) and Velvet (7). Exonerate servers will be used for searching all assembled sequences. We have extended Velvet, a de Bruijn graph-based sequence assembler, to support sequence similarity searches against assemblies induced from trace and short read sequences. We have implemented Velvet as a server that uses the Exonerate client server protocol so that we can run the Exonerate client against both Exonerate and Velvet servers. We have extended the exonerate client to support multiple and redundant servers to maximize the availability of our sequence search service. The result for the user will be a simple search page from which searches across comprehensive data can be launched, using Exonerate or Velvet methods as appropriate according to the nature of the data to be searched.
Presently, sequence similarity searches for ENA data are available using web, as well as EBI SOAP and REST Web Services interfaces (8). Search against ENA-Annotation sequences is available using NCBI-Blast (9) at http://www.ebi.ac.uk/Tools/sss/ncbiblast/nucleotide.html and Fasta (10) at http://www.ebi.ac.uk/Tools/sss/fasta/nucleotide.html. WGS sequences and full genomes are available for Fasta search at http://www.ebi.ac.uk/Tools/sss/fasta/wgs.html and http://www.ebi.ac.uk/Tools/sss/fasta/genomes.html, respectively.
FUNDING
Funding for open access charge: European Molecular Biology Laboratory and the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 2.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansorge WJ. Next-generation DNA sequencing techniques. N. Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugawara H, Ikeo K, Fukuchi S, Gojobori T, Tateno Y. DDBJ dealing with mass data produced by the second generation sequencer. Nucleic Acids Res. 2009;37:D16–D18. doi: 10.1093/nar/gkn724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slater G, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McWilliam H, Valentin F, Goujon M, Li W, Narayanasamy M, Martin J, Miyar T, Lopez R. Web services at the European Bioinformatics Institute. Nucleic Acids Res. 2009;37:W6–W10. doi: 10.1093/nar/gkp302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. PNAS. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]