Abstract
The Encyclopedia of DNA Elements (ENCODE) project is an international consortium of investigators funded to analyze the human genome with the goal of producing a comprehensive catalog of functional elements. The ENCODE Data Coordination Center at The University of California, Santa Cruz (UCSC) is the primary repository for experimental results generated by ENCODE investigators. These results are captured in the UCSC Genome Bioinformatics database and download server for visualization and data mining via the UCSC Genome Browser and companion tools (Rhead et al. The UCSC Genome Browser Database: update 2010, in this issue). The ENCODE web portal at UCSC (http://encodeproject.org or http://genome.ucsc.edu/ENCODE) provides information about the ENCODE data and convenient links for access.
BACKGROUND
With the completion of the draft sequence of the human genome in 2003, the ENCODE project (http://www.genome.gov/ENCODE) (1) was initiated as a follow-on project focused on identifying functional elements in the genome using a variety of experimental methods.
ENCODE pilot phase
ENCODE began as a pilot project focusing on 1% of the human genome. Results from this phase of ENCODE were reported in Nature (2) and a special issue of Genome Biology in June 2007 (3).
Data from this phase are available at UCSC in designated ENCODE ‘track groups’ within the UCSC browsers for the hg16, hg17 and hg18 human genome assemblies (NCBI Builds 34–36) (4–6). The pilot section of the UCSC ENCODE web portal (http://genome.ucsc.edu/ENCODE/pilot.html) supplies information about this phase of ENCODE, and a ‘Regions’ link on this page (http://genome.ucsc.edu/ENCODE/encode.hg18.html) provides convenient access to the areas of the genome with ENCODE pilot phase annotations.
ENCODE production (scale-up) phase
In September 2007, the ENCODE project scaled up to production mode, with the goal of generating high-throughput annotations on the full human genome. In addition to the increased scale and data volume, other aspects of the project expanded in an effort to standardize results and facilitate integrative analysis. Significant differences from the pilot phase include:
Common cell types (http://www.genome.gov/26524238) and approved cell culture protocols
Specification of standards for experiment verification and reporting
Capture of experiment metadata using controlled vocabularies
New experimental technologies based on high-throughput sequencing
A data release policy restricting use of data for nine months following release
To accommodate the increased scale and volume of ENCODE data submissions, the ENCODE project at UCSC was expanded to include a more formal data submission process with substantial automation. The browser and download sites were expanded to include new data types, the capture of additional metadata, and new track organization features (described below).
Related projects
In parallel with the ENCODE project, the modENCODE project (http://www.modencode.org/) (7) aims to similarly study the genomes of two model organisms: worm (Caenorhabditis elegans) and fruitfly (Drosophila melanogaster).
ENCODE DATA AT UCSC
As of September 2009, the ENCODE DCC has processed a full year of production-phase data submissions from the ENCODE data providers, representing four defined data freezes (Nov08, Feb09, Jul09 and Sep09). A total of 341 experiments have been submitted to the DCC, and 207 of these—in 18 browser tracks—have been released to the UCSC public server after quality review. These tracks include chromatin immunoprecipitation experiments for transcription factor binding and histone modification; maps of open chromatin, chromatin interactions, and DNA methylation; transcriptome profiling of whole cell and cellular compartments by RNA-seq and microarray; and identification of transcript ends together with high-quality gene annotations.
The goal of the initial ENCODE freezes was to provide a comprehensive matrix of experiment results in two common cell lines—K562 leukemia and GM12878 lymphoblastoid (a 1000 genomes deep-sequence sample). The ENCODE Consortium defined these two cell lines as ‘Tier1’, required for use by all ENCODE groups. This standardization ensures greater consistency between different tracks. An additional five cell types (HeLaS3, HepG2, NHEK, HUVEC and H1ES) were designated ‘Tier2’, shared by many groups. Finally, individual labs have registered for use an additional 68 cell types designated ‘Tier3’. The full list of cell types in use by ENCODE, with vendor IDs and cell culture protocol documentation, is available from the ‘Cell Types’ link at the UCSC ENCODE portal (http://genome.ucsc.edu/ENCODE/cellTypes.html).
For each experiment type (ChIP-seq, DNase-seq, etc.), the ENCODE investigators conduct multiple experiments, using different cell lines, tissue samples and (as appropriate) other variables for the experiment type. Transcriptome experiments typically vary the RNA extracts (e.g. polyA+, polyA−, total or short) and the subcellular compartment from which the extract was obtained (e.g. nucleus, cytosol, nucleolus or whole cell). Chromatin immunoprecipitation to localize transcription factor binding or regions of histone marks is performed with differing antibodies. ENCODE investigators have registered 59 antibodies with the DCC.
Table 1 summarizes the experiments submitted to the ENCODE DCC as of mid-September 2009. See the ‘Data submission status spreadsheet’ (Supplementary Data S1) for a complete list of submitted experiments with status.
Table 1.
Summary of ENCODE datasets, as of 15 September 2009
| Data type | Description | Investigators | Number of experiments |
|---|---|---|---|
| BiP | Bi-directional promoters | NHGRI | 2 |
| CAGE | 5′ cap analysis gene expression | Riken | 11 |
| ChIP-seq | TF and polymerase binding, histone marks by ChIP | Yale, UC Davis, HudsonAlpha, Broad, UW, UNC | 185 |
| DNA-seq | DNA fragment sequencing | Genome Inst Singapore | 5 |
| DNase-seq | DNaseI hypersensitivity | UW, Duke | 20 |
| Exon-array | Gene expression by all-exon microarray | Affymetrix/CSHL | 10 |
| FAIRE-seq | Formaldehyde Assisted Isolation of Regulatory Elements | U. Texas | 5 |
| Genes | High-quality gene annotations | Gencode/Sanger | 3 |
| Mapability | Uniqueness of short read nmers | Broad, Duke, UMass | 5 |
| Methyl27 | DNA methylation by Illumina 27K | HudsonAlpha | 3 |
| Methyl-seq | DNA methylation by restriction enzymes | HudsonAlpha | 15 |
| NRE | Negative regulatory elements | NHGRI | 6 |
| PET | 5′- and 3′-paired-end tags | Genome Inst. Singapore | 13 |
| RIP-chip | RNA-binding proteins | SUNY Albany | 7 |
| RNA-chip | RNA microarray | Affymetrix/CSHL | 25 |
| RNA-seq | RNA sequencing | Caltech, CSHL, GIS, Yale | 23 |
| TbaAlign | Multi-species alignment with TBA | NHGRI | 1 |
| CNV | Copy number variation | HudsonAlpha | 3 |
| DHS-5C | Chromatin interactions: DHS versus TSS | U Washington | 2 |
| 5C | Chromatin interactions: pilot region | U Mass | 2 |
| Total | 341 |
The ENCODE Consortium has made a major effort to standardize experimental methods, analysis strategies and data reporting protocols. During the transition from pilot to production phase, the bulk of ENCODE investigators shifted methodologies from microarray to assays based on short read sequencing technologies including ChIP-seq, DNase-seq, RNA-seq and Methyl-seq. The DCC has been active in developing file formats, database designs and browser track displays to accommodate these new data types. The ‘Sample ENCODE Session’ in the Supplementary Data S2 provides a Genome Browser screen shot showing a broad sampling of ENCODE data.
ACCESSING THE ENCODE DATA
UCSC provides three major methods of accessing the ENCODE data. For viewing multiple ENCODE experiments simultaneously alongside standard annotations such as gene positions, the Genome Browser is the method of choice. The Genome Browser displays the data graphically and works well on regions of up to tens of megabases in size. The Table Browser provides access to the same data in a variety of easily parseable formats, offering basic but useful data analysis as well such as the ability to compute intersections and correlations between tracks. The Table Browser interface parallels that of the Genome Browser, which facilitates finding the data tables that correspond to a particular track. Finally, all ENCODE data are available as downloadable files on the UCSC FTP site.
In general, we recommend getting familiar with the data graphically in the Genome Browser first, then using the Table Browser to explore the organization of the database and to download subsets of data no larger than a chromosome. For access to full-genome data, it is best to download the data as files from the FTP site. ENCODE tracks are standard tracks in the UCSC genome database; therefore, all tools available at the site can be applied to ENCODE data.
Visualizing data in the genome browser
Whole-genome ENCODE data generated during the ENCODE production phase are loaded into the standard browser track groups in the UCSC genome database (in contrast to pilot phase data, which were placed in ENCODE-specific groups). Nearly all of the ENCODE data can be found in the ‘expression’ and ‘regulation’ track groups; a few ENCODE tracks are located in the ‘mapping’, ‘genes’ and ‘variation’ groups. ENCODE tracks are highlighted in the browser track menus by an NHGRI helix logo (Figure 1). The ‘Release Log’ link at the UCSC ENCODE portal (http://genome.ucsc.edu/ENCODE/releaseLog.html) provides access to the list of released ENCODE tracks, along with links to the methods description and configuration for each track.
Figure 1.
A portion of the Genome Browser track group controls section on the hg18 human assembly, showing tracks in the ‘expression’ and ‘regulation’ track groups. The ENCODE tracks are distinguished by the NHGRI helix icon appearing in the label.
To make the hundreds of ENCODE tracks more manageable for users, we have enhanced the UCSC Genome Browser track configuration to provide more power, flexibility and interactivity. Subtracks can now be individually customized, organized into multiple ‘views’, and reordered by column sort or by drag-and-drop. We have incorporated a structured metadata display on Genome Browser track details pages and have added a link to facilitate bulk download of data files associated with a track.
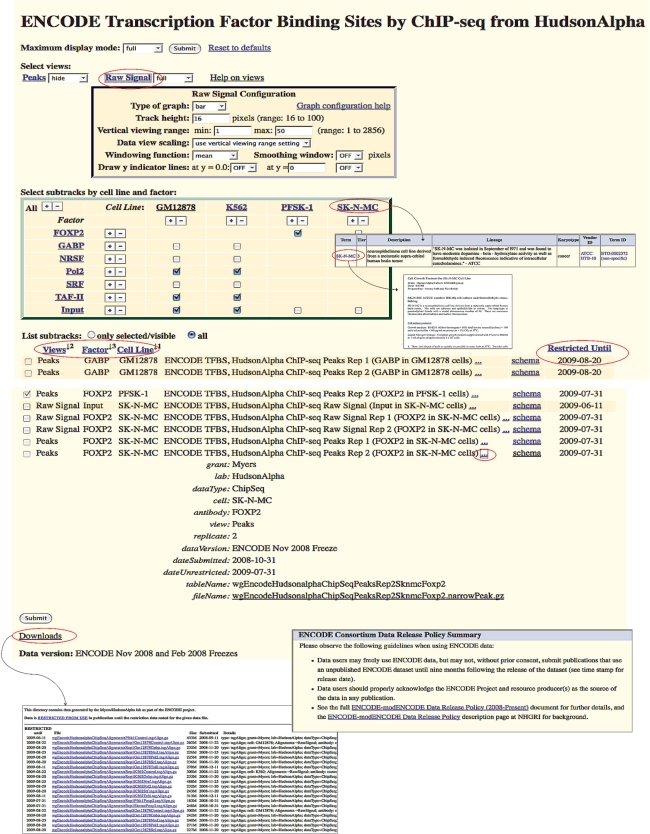
Figure 2 provides a detailed look at these new features. The ‘Views’ section near the top of the track configuration page shows the potentially multiple data representations for a single experiment. Efforts have been made to standardize ‘views’ across similar datasets in ENCODE. Most tracks follow one of two patterns:
Regulatory elements: Peaks (discrete sites) and Signal (continuous graph of enrichment)
Gene expression: Plus and Minus Signal (coverage graph of reads on forward and reverse strand) and Alignments (short reads aligned to genome)
Below the ‘Views’ section, configuration pages for ENCODE tracks typically include a matrix of checkboxes that allow the selection of subtracks by experimental variables such as cell type or antibody. Subtracks can also be selected individually from the list of all subtracks displayed at the bottom of the configuration section. The column headers of this section (which include the experimental variables shown in the matrix) define the ordering of subtracks within the track display. The subtrack ordering can be changed by clicking the column headers to reorder by group, or by dragging and dropping individual subtracks in the list.
Figure 2.
Example configuration and details pages for an ENCODE track, showing important navigation and informational items.
The clickable (…) icons expand the display to show the metadata (experiment type and variables, data format and data freeze) for each subtrack. Clicking the ‘schema’ link for any subtrack listed on the track configuration page displays a full description of the data representation. The database representations and file formats for the peaks and alignments data were designed specifically for ENCODE. Signal views use one of the standard UCSC graphing formats: wiggle, bedGraph or bigWig.
Finally, note the ‘restricted until’ date for each subtrack, which shows the date when restricted use of the data expires. The data use policy for ENCODE is described in more detail below.
Bulk downloads of data
The DCC provides both raw data (sequence reads and quality scores) and processed data files (alignments, density graphs and peak calls). The raw data from high-throughput sequencing are provided in FASTQ format when feasible. SOLID colorspace sequences and quality are provided in CSFASTA and CSQUAL format.
ENCODE files can be retrieved by web access or anonymous FTP from the UCSC download server. Due to the large size of most ENCODE data sets, FTP retrieval is recommended.
The ENCODE portal includes a Downloads index page (http://genome.ucsc.edu/ENCODE/downloads.html) that provides convenient web access to data files by track. The top-level download area for ENCODE data is at http://hgdownload.cse.ucsc.edu/goldenPath/hg18/encodeDCC.
For FTP access, connect to the FTP server at ‘hgdownload.cse.ucsc.edu’, then move to the ‘goldenPath/hg18/encodeDCC’ directory. Each of the listed subdirectories contains the data files for an individual ENCODE track (one track for each data type per lab), along with an index.html page listing the data files, metadata describing the experiment, the type, experimental variables, the data format and a data restriction timestamp. An example is shown in Figure 2.
For convenient access to the ENCODE data in the Genome Browser, a Downloads link is included on the track configuration page below the subtrack selection list.
Data use policy
The following guidelines should be followed when using ENCODE data:
Data users may freely use ENCODE data, but may not, without prior consent, submit publications that use an unpublished ENCODE dataset until nine months following the release of the dataset (see time stamp for release date).
Data users should properly acknowledge the ENCODE Project and resource producer(s) as the source of the data in any publication.
See the full ENCODE Data Release Policy (2008–present) document (http://www.genome.gov/Pages/Research/ENCODE/ENCODEDataReleasePolicyFinal2008.pdf) for further details.
Outreach and tutorials
Additional informational materials, including free tutorials describing access to the ENCODE data and use of the UCSC Genome Browser, are available from OpenHelix at http://www.openhelix.com/.
FUTURE DIRECTIONS
HG19 (GRCh37) human genome assembly
As of September 2009, all ENCODE results for the production phase of ENCODE have been reported on the hg18 (NCBI Build 36) genome assembly. The ENCODE Consortium plans to migrate to the newer human genome assembly in late 2009 or early 2010. As part of the migration, the DCC will convert the coordinates on annotations produced in the initial years of the project to the new assembly.
Mouse genome
The ENCODE project plans to expand to include the study of the Mus musculus genome beginning in late 2009.
Track search tool
The breadth of ENCODE data creates a challenge in terms of presentation—how to provide access to the full range of data without overwhelming the user? The extension of the existing track organization mechanisms to provide a hierarchy of data (i.e. multiview) improves on a linear listing of thousands of datasets and files. To further facilitate the dataset selection process, UCSC is planning to develop a more intuitive track search mechanism that supports the entry of keywords indicating the type of data desired.
RNA-seq display and file formats
As the technology for transcriptome profiling advances, with longer read lengths, paired reads and mapping across splice junctions, a richer data representation and browser display is called for. Binary Alignment/Map (BAM) format is a binary representation of the Sequence Alignment/Map (SAM) format developed for the 1000 Genomes Project (8). SAM/BAM provides a rich, efficient and standard method of capturing sequence alignments from high-throughput sequencing in a platform-independent manner. UCSC has implemented a browser display for BAM files, which we plan to include as a supported ENCODE data format in the coming year.
CONTACTING US
Questions and feedback about the ENCODE data at UCSC should be directed to our ENCODE mailing list: encode@soe.ucsc.edu. General questions about the Genome Browser should be sent to the mailing lists described in the Genome Browser companion paper in this issue. We announce releases of new ENCODE data via the ENCODE announcement list, encode-announce@soe.ucsc.edu; to subscribe, visit https://lists.soe.ucsc.edu/mailman/listinfo/encode-announce.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Human Genome Research Institute (5P41HG002371-09 to the UCSC Center for Genomic Science and 5U41HG004568-02 to the UCSC ENCODE Data Coordination Center); Howard Hughes Medical Institute (to D.H.). T.W. is a Helen Hay Whitney fellow. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. K.R.R., T.R.D., M.P., G.P.B., L.R.M., A.P., B.J.R., A.S.H., A.S.Z., B.R., K.E.S., P.A.F., R.M.K., D.K., D.H. and W.J.K. receive royalties from the sale of UCSC Genome Browser source code licenses to commercial entities.
ACKNOWLEDGEMENTS
We thank the members of the ENCODE Consortium for their collaborative spirit and stamina over the six years of data production, submission and analysis that the ENCODE project has required to date. We also acknowledge Hiram Clawson, a core Genome Browser engineer who has contributed greatly to its overall success by his work to keep the browser reliable, fast and annotation-rich. We thank the UCSC CCDS team, Mark Diekhans and Rachel Harte, for their contributions to the Gencode genes and their tireless advocacy for the best data representations and display for the challenging and high-value RNA-seq data. Nicole Washington and Lincoln Stein at the modENCODE DCC have graciously shared DCC processes and strategies. Melissa Cline provided technical review and editing for this paper, for which we thank her. And finally, we acknowledge our dedicated team of system administrators, Jorge Garcia, Erich Weiler, Victoria Lin and Alex Wolfe, for their relentless provision of more cycles and megabytes, valiant swat-team trouble-shooting and for generally providing an outstanding computing environment.
REFERENCES
- 1.ENCODE Consortium. The ENCODE (ENCyclopedia Of DNA Elements) project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 2.The ENCODE Project Consortium. Birney E, Stamatoyannopoulos J, Dutta A, Guigó R, Gingeras T, Margulies E, Weng Z, Snyder M, Dermitzakis E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinstock GM. ENCODE: more genomic empowerment. Genome Res. 2007;17:667–668. doi: 10.1101/gr.6534207. [DOI] [PubMed] [Google Scholar]
- 4.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith K, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas DJ, Rosenbloom KR, Clawson H, Hinrichs AS, Trumbower H, Raney BJ, Karolchik D, Barber GP, Harte RA, Hillman-Jackson J, et al. The ENCODE project at UC Santa Cruz. Nucleic Acids Res. 2007;35:D663–D667. doi: 10.1093/nar/gkl1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celniker S, Dillon L, Gerstein M, Gunsalus K, Henikoff S, Karpen G, Kellis M, Lai E, Lieb J, MacAlpine D, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map (SAM) Format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]